Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1067-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1067-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 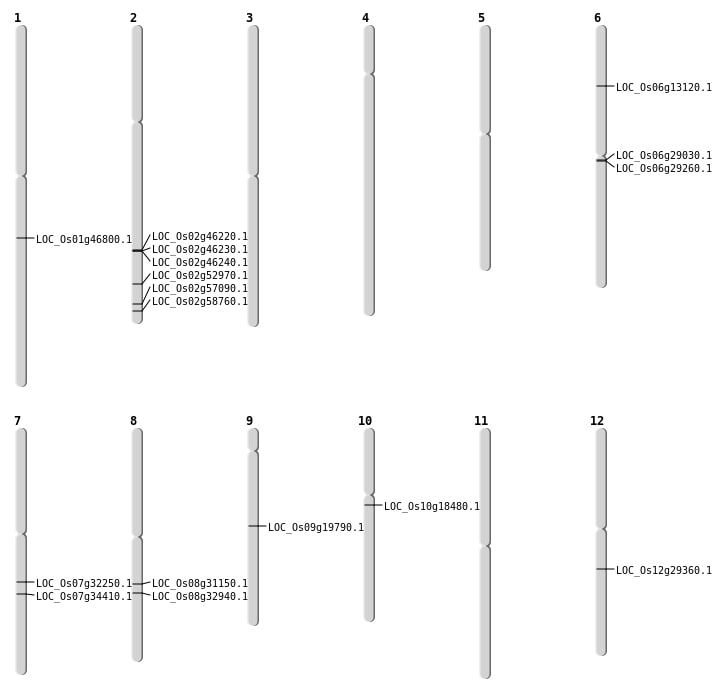
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 5127223 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 921844 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 12206886 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 9385786 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g18480 |
| SBS | Chr11 | 11871483 | C-A | HET | INTRON | |
| SBS | Chr11 | 14254430 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 17433316 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g29360 |
| SBS | Chr12 | 8233157 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 28377111 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 30277452 | G-T | HET | INTRON | |
| SBS | Chr2 | 30594249 | A-C | HET | INTRON | |
| SBS | Chr2 | 32409771 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g52970 |
| SBS | Chr3 | 20852952 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 28705585 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 3546013 | C-T | HET | INTRON | |
| SBS | Chr4 | 12405400 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 16226138 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 34563303 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 25289523 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 27929428 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 6176885 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 6516659 | A-C | HET | UTR_5_PRIME | |
| SBS | Chr7 | 16854456 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 19167801 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g32250 |
| SBS | Chr7 | 6894587 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 9888677 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 19254022 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g31150 |
| SBS | Chr9 | 13023124 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr9 | 13180443 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 4317486 | C-G | HOMO | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 7582134 | 7582137 | 3 | |
| Deletion | Chr1 | 8329412 | 8329413 | 1 | |
| Deletion | Chr5 | 8443589 | 8443590 | 1 | |
| Deletion | Chr5 | 8686860 | 8686866 | 6 | |
| Deletion | Chr2 | 9039366 | 9039368 | 2 | |
| Deletion | Chr6 | 11141756 | 11141761 | 5 | |
| Deletion | Chr9 | 11843884 | 11843886 | 2 | LOC_Os09g19790 |
| Deletion | Chr2 | 12661183 | 12661315 | 132 | |
| Deletion | Chr11 | 14130080 | 14130105 | 25 | |
| Deletion | Chr6 | 15146569 | 15146577 | 8 | |
| Deletion | Chr6 | 16544658 | 16548907 | 4249 | LOC_Os06g29030 |
| Deletion | Chr6 | 16715367 | 16715370 | 3 | LOC_Os06g29260 |
| Deletion | Chr3 | 16895990 | 16896009 | 19 | |
| Deletion | Chr9 | 18458778 | 18458788 | 10 | |
| Deletion | Chr7 | 18518651 | 18518657 | 6 | |
| Deletion | Chr8 | 19254016 | 19254017 | 1 | LOC_Os08g31150 |
| Deletion | Chr1 | 26688232 | 26688233 | 1 | LOC_Os01g46800 |
| Deletion | Chr6 | 27196718 | 27196719 | 1 | |
| Deletion | Chr2 | 28162001 | 28200000 | 38000 | 3 |
| Deletion | Chr3 | 29298489 | 29298500 | 11 | |
| Deletion | Chr1 | 30131824 | 30131830 | 6 | |
| Deletion | Chr2 | 34946303 | 34946313 | 10 | |
| Deletion | Chr2 | 34958483 | 34958484 | 1 | LOC_Os02g57090 |
| Deletion | Chr2 | 35897689 | 35897693 | 4 | LOC_Os02g58760 |
| Deletion | Chr1 | 37144851 | 37144854 | 3 |
Insertions: 5
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 3330019 | 7857443 | |
| Inversion | Chr2 | 12661184 | 28199842 | LOC_Os02g46240 |
| Inversion | Chr12 | 15175975 | 16534897 | |
| Inversion | Chr8 | 20430809 | 20879680 | LOC_Os08g32940 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 7596518 | Chr7 | 13502633 | |
| Translocation | Chr10 | 18419178 | Chr6 | 7203363 | LOC_Os06g13120 |
| Translocation | Chr10 | 18419181 | Chr6 | 7191921 | |
| Translocation | Chr7 | 20641689 | Chr2 | 28399330 | LOC_Os07g34410 |
