Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1069-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1069-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 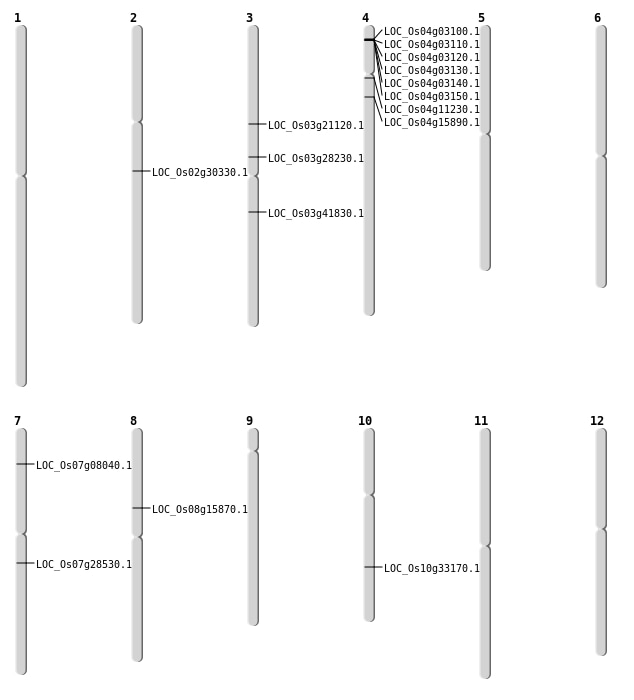
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14158371 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 41746721 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 495848 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 9947928 | C-A | HET | UTR_5_PRIME | |
| SBS | Chr10 | 11138892 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 4287227 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 12973844 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 21084573 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 25168808 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 5578972 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 15613360 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 18437345 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 4304871 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 4828594 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 8187803 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 9630704 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 8443748 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 14140204 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 16241562 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g28230 |
| SBS | Chr3 | 23965818 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 27626922 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 15515901 | G-A | HET | INTRON | |
| SBS | Chr4 | 16182014 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 8627164 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g15890 |
| SBS | Chr5 | 1804760 | C-G | HET | INTRON | |
| SBS | Chr5 | 5159719 | A-G | HET | INTRON | |
| SBS | Chr6 | 24849246 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 14435583 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 16697957 | C-A | HOMO | STOP_GAINED | LOC_Os07g28530 |
| SBS | Chr7 | 7844806 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 9804660 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 9650377 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g15870 |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1278001 | 1315000 | 37000 | 6 |
| Deletion | Chr10 | 2233534 | 2233565 | 31 | |
| Deletion | Chr11 | 2744952 | 2744953 | 1 | |
| Deletion | Chr6 | 4790293 | 4790296 | 3 | |
| Deletion | Chr4 | 5671722 | 5671723 | 1 | |
| Deletion | Chr11 | 6067463 | 6067467 | 4 | |
| Deletion | Chr11 | 7019074 | 7019075 | 1 | |
| Deletion | Chr1 | 7853347 | 7853357 | 10 | |
| Deletion | Chr12 | 8022081 | 8022098 | 17 | |
| Deletion | Chr12 | 10779629 | 10779634 | 5 | |
| Deletion | Chr4 | 11551981 | 11551982 | 1 | |
| Deletion | Chr5 | 12850211 | 12850212 | 1 | |
| Deletion | Chr5 | 13769197 | 13769207 | 10 | |
| Deletion | Chr4 | 14875074 | 14875076 | 2 | |
| Deletion | Chr12 | 16933899 | 16933908 | 9 | |
| Deletion | Chr4 | 16958751 | 16958765 | 14 | |
| Deletion | Chr4 | 17102396 | 17102397 | 1 | |
| Deletion | Chr2 | 18069617 | 18069665 | 48 | LOC_Os02g30330 |
| Deletion | Chr8 | 20892862 | 20892898 | 36 | |
| Deletion | Chr4 | 22441702 | 22441705 | 3 | |
| Deletion | Chr3 | 23228421 | 23228422 | 1 | LOC_Os03g41830 |
| Deletion | Chr5 | 23649560 | 23649573 | 13 | |
| Deletion | Chr11 | 27696316 | 27696318 | 2 | |
| Deletion | Chr3 | 30498804 | 30498806 | 2 | |
| Deletion | Chr3 | 33928073 | 33928082 | 9 | |
| Deletion | Chr1 | 35522096 | 35522097 | 1 |
Insertions: 8
No Inversion
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 6139703 | Chr1 | 17190459 | LOC_Os04g11230 |
| Translocation | Chr11 | 13215626 | Chr3 | 12026781 | LOC_Os03g21120 |
| Translocation | Chr11 | 13217136 | Chr3 | 12026830 | LOC_Os03g21120 |
| Translocation | Chr10 | 17375331 | Chr7 | 4074187 | 2 |
| Translocation | Chr10 | 17375336 | Chr7 | 4074198 | 2 |
