Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1086-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1086-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 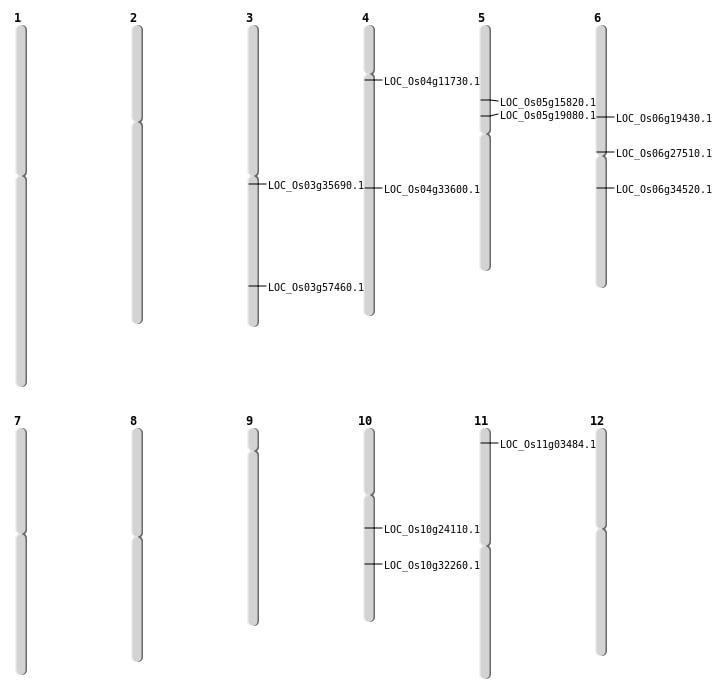
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10124854 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 10618104 | T-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 18517171 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 25557358 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 32335254 | T-C | HET | INTRON | |
| SBS | Chr1 | 3555044 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 38985634 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 5155659 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 5263225 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 5658829 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 12355755 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g24110 |
| SBS | Chr10 | 15217677 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 16935003 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g32260 |
| SBS | Chr10 | 2724482 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 1336038 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g03484 |
| SBS | Chr11 | 13499617 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 19299323 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 10753975 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 23536198 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 20161171 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 24218704 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 22420180 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 268419 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 33511067 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 17657890 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 2240496 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 2240555 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 4168906 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 5147850 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 11069218 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g19430 |
| SBS | Chr6 | 14022592 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 15574010 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g27510 |
| SBS | Chr6 | 20086297 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g34520 |
| SBS | Chr6 | 6494224 | A-G | HET | INTRON | |
| SBS | Chr6 | 7549286 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 14910713 | A-C | HOMO | INTRON | |
| SBS | Chr8 | 12190858 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 14571144 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 9688631 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 17382564 | A-T | HOMO | INTRON | |
| SBS | Chr9 | 18052310 | A-C | HOMO | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1969728 | 1969729 | 1 | |
| Deletion | Chr8 | 2626546 | 2626548 | 2 | |
| Deletion | Chr10 | 3724761 | 3724763 | 2 | |
| Deletion | Chr1 | 5711031 | 5711033 | 2 | |
| Deletion | Chr4 | 6429258 | 6429261 | 3 | LOC_Os04g11730 |
| Deletion | Chr1 | 6798220 | 6798229 | 9 | |
| Deletion | Chr5 | 7628140 | 7628141 | 1 | |
| Deletion | Chr5 | 9043531 | 9043535 | 4 | |
| Deletion | Chr12 | 9733441 | 9733443 | 2 | |
| Deletion | Chr10 | 12158976 | 12158978 | 2 | |
| Deletion | Chr8 | 12782747 | 12782753 | 6 | |
| Deletion | Chr10 | 16077343 | 16077352 | 9 | |
| Deletion | Chr10 | 17869340 | 17869369 | 29 | |
| Deletion | Chr9 | 18097825 | 18097826 | 1 | |
| Deletion | Chr3 | 19767557 | 19767565 | 8 | LOC_Os03g35690 |
| Deletion | Chr6 | 22677457 | 22677464 | 7 | |
| Deletion | Chr8 | 23277604 | 23277637 | 33 | |
| Deletion | Chr3 | 32760611 | 32760613 | 2 | LOC_Os03g57460 |
| Deletion | Chr1 | 38708711 | 38708717 | 6 |
Insertions: 5
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 8935119 | 9334720 | LOC_Os05g15820 |
| Inversion | Chr5 | 9334721 | 11075627 | LOC_Os05g19080 |
| Inversion | Chr4 | 20348609 | 20532269 | LOC_Os04g33600 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 1881833 | Chr2 | 28621085 | |
| Translocation | Chr4 | 20348615 | Chr3 | 18647676 | LOC_Os04g33600 |
| Translocation | Chr4 | 20532276 | Chr3 | 18648029 |
