Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1089-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1089-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 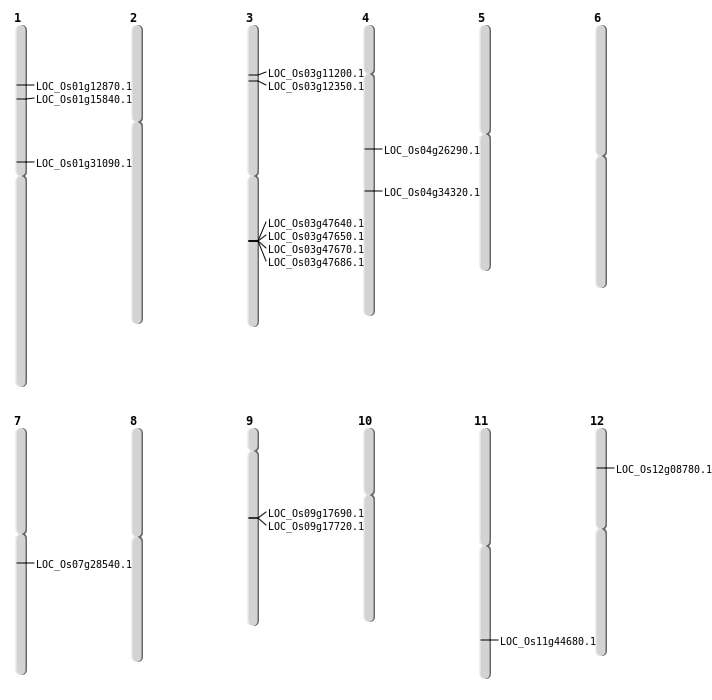
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16999007 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g31090 |
| SBS | Chr1 | 20286934 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 20927405 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 7127325 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g12870 |
| SBS | Chr10 | 11029918 | A-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 20874999 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 18415974 | A-G | HET | INTRON | |
| SBS | Chr11 | 25524108 | G-A | HET | INTRON | |
| SBS | Chr11 | 27019946 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g44680 |
| SBS | Chr12 | 14186464 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 18938844 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr12 | 20996112 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 24571455 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 4417373 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 11385050 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 22867403 | T-C | HET | INTRON | |
| SBS | Chr2 | 23194480 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 30423723 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 7212026 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 11392487 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 2773955 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 28662391 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 3763679 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 15325223 | C-A | HOMO | STOP_GAINED | LOC_Os04g26290 |
| SBS | Chr4 | 19045630 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 20786979 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g34320 |
| SBS | Chr4 | 26340740 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 20369899 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 4840549 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 5585103 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 10625428 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 20211010 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 25778143 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 29134383 | C-G | HET | UTR_3_PRIME | |
| SBS | Chr6 | 8401859 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 16703086 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 18745419 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 22579352 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 26826378 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 26038646 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5925673 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 14361287 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 7504043 | C-T | HET | INTERGENIC |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 395411 | 395431 | 20 | |
| Deletion | Chr11 | 1584476 | 1584477 | 1 | |
| Deletion | Chr2 | 1799322 | 1799330 | 8 | |
| Deletion | Chr2 | 2130991 | 2131023 | 32 | |
| Deletion | Chr6 | 2139657 | 2139663 | 6 | |
| Deletion | Chr12 | 4517276 | 4517288 | 12 | LOC_Os12g08780 |
| Deletion | Chr4 | 5067879 | 5067906 | 27 | |
| Deletion | Chr3 | 5716271 | 5716272 | 1 | |
| Deletion | Chr7 | 9201688 | 9201701 | 13 | |
| Deletion | Chr11 | 11643019 | 11643030 | 11 | |
| Deletion | Chr8 | 12838481 | 12838485 | 4 | |
| Deletion | Chr9 | 14379177 | 14379179 | 2 | |
| Deletion | Chr7 | 16700384 | 16700476 | 92 | LOC_Os07g28540 |
| Deletion | Chr1 | 20887058 | 20887066 | 8 | |
| Deletion | Chr12 | 24675304 | 24675308 | 4 | |
| Deletion | Chr3 | 26984001 | 27021000 | 37000 | 4 |
| Deletion | Chr3 | 26986104 | 27020899 | 34795 | 3 |
| Deletion | Chr1 | 42748264 | 42748685 | 421 |
Insertions: 8
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 5766949 | 6515542 | 2 |
| Inversion | Chr1 | 8923533 | 9732266 | LOC_Os01g15840 |
| Inversion | Chr9 | 9961390 | 10821850 | LOC_Os09g17690 |
| Inversion | Chr9 | 10821876 | 10833833 | 2 |
| Inversion | Chr6 | 14464932 | 14524940 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 6747445 | Chr8 | 19016788 |
