Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1103-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1103-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 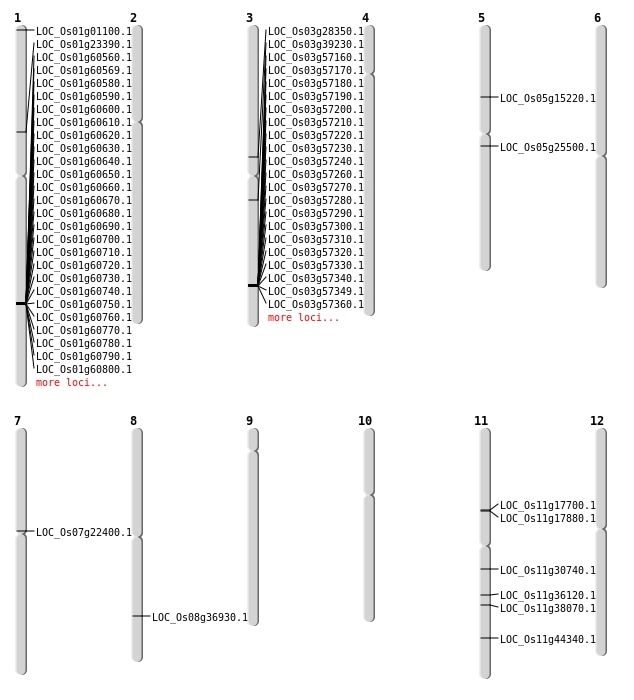
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13145221 | G-A | HET | START_LOST | LOC_Os01g23390 |
| SBS | Chr1 | 16515155 | T-G | HET | INTRON | |
| SBS | Chr1 | 21831597 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 23332079 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 24176684 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 24176686 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 27100579 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 34545523 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 36317936 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g62720 |
| SBS | Chr1 | 39165388 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr1 | 52129 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g01100 |
| SBS | Chr11 | 17898471 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g30740 |
| SBS | Chr11 | 21233703 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g36120 |
| SBS | Chr12 | 12824701 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 15395602 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 19993687 | T-A | HET | INTRON | |
| SBS | Chr12 | 4962219 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 1183229 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 32965525 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 32965757 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr3 | 12000961 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 14540438 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 16331284 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g28350 |
| SBS | Chr3 | 2553157 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 33481823 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 33481828 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 36076155 | C-T | HET | INTRON | |
| SBS | Chr3 | 7406207 | T-C | HET | INTRON | |
| SBS | Chr4 | 13911085 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 21091677 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 26280030 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 13437460 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 9971806 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 12575890 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g22400 |
| SBS | Chr7 | 26221605 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 3145563 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 23367494 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g36930 |
| SBS | Chr8 | 623306 | A-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 2877462 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 7105022 | A-T | HET | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 2509688 | 2509696 | 8 | |
| Deletion | Chr4 | 4977620 | 4977688 | 68 | |
| Deletion | Chr6 | 4987718 | 4987719 | 1 | |
| Deletion | Chr5 | 8635484 | 8635485 | 1 | LOC_Os05g15220 |
| Deletion | Chr5 | 8933206 | 8933207 | 1 | |
| Deletion | Chr6 | 13032360 | 13032364 | 4 | |
| Deletion | Chr5 | 14825427 | 14825428 | 1 | LOC_Os05g25500 |
| Deletion | Chr10 | 16400989 | 16400991 | 2 | |
| Deletion | Chr2 | 17514739 | 17514745 | 6 | |
| Deletion | Chr10 | 17652901 | 17652905 | 4 | |
| Deletion | Chr5 | 18706226 | 18706251 | 25 | |
| Deletion | Chr3 | 19823819 | 19823939 | 120 | |
| Deletion | Chr8 | 20069865 | 20069871 | 6 | |
| Deletion | Chr3 | 21174861 | 21174863 | 2 | |
| Deletion | Chr9 | 21247119 | 21247121 | 2 | |
| Deletion | Chr3 | 21571647 | 21571651 | 4 | |
| Deletion | Chr3 | 21785893 | 21785900 | 7 | LOC_Os03g39230 |
| Deletion | Chr9 | 22088474 | 22088475 | 1 | |
| Deletion | Chr3 | 24069308 | 24069315 | 7 | |
| Deletion | Chr11 | 24878806 | 24878807 | 1 | |
| Deletion | Chr11 | 26802247 | 26802248 | 1 | LOC_Os11g44340 |
| Deletion | Chr8 | 27085374 | 27085388 | 14 | |
| Deletion | Chr8 | 27604940 | 27604948 | 8 | |
| Deletion | Chr2 | 29715039 | 29715043 | 4 | |
| Deletion | Chr3 | 32598179 | 32784108 | 185929 | 34 |
| Deletion | Chr1 | 35017001 | 35212000 | 195000 | 32 |
| Deletion | Chr1 | 39236123 | 39236125 | 2 |
Insertions: 5
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 9878540 | 10014775 | 2 |
| Inversion | Chr11 | 9878554 | 10014784 | 2 |
| Inversion | Chr1 | 34758217 | 35127612 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 22578753 | Chr8 | 3787816 | LOC_Os11g38070 |
