Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1108-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1108-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 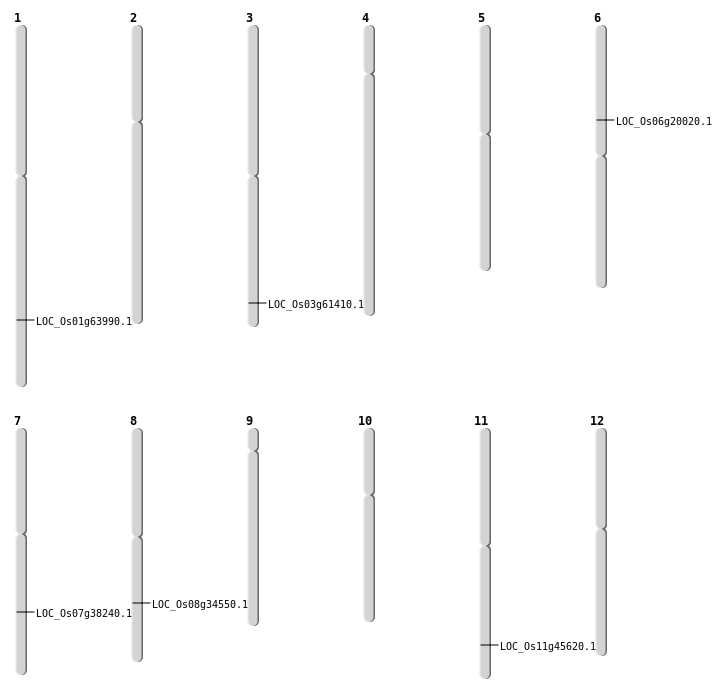
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11996101 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 15309924 | C-T | HET | INTRON | |
| SBS | Chr1 | 35505778 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr1 | 37497191 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 42468013 | C-A | HET | INTRON | |
| SBS | Chr1 | 5070315 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 9470130 | T-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 11543382 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr10 | 13768800 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 15500137 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 9973230 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 15184707 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 24763424 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 521210 | C-A | HET | INTRON | |
| SBS | Chr11 | 6002826 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 6449964 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 8849387 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 12610745 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 14831299 | G-C | HET | INTERGENIC | |
| SBS | Chr12 | 1820014 | C-T | HET | INTRON | |
| SBS | Chr12 | 23544788 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 11608366 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 24121099 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 699997 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 11847648 | T-C | HOMO | INTRON | |
| SBS | Chr3 | 14136193 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 21370465 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 28984926 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 14547660 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 22427400 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 2793955 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 3193149 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 11332436 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 13687749 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 16404526 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 18788689 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 4659133 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 11477729 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g20020 |
| SBS | Chr6 | 12709952 | A-T | HOMO | INTRON | |
| SBS | Chr7 | 10292781 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 11253352 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 13883208 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 20466247 | T-A | HOMO | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 3035336 | 3035338 | 2 | |
| Deletion | Chr2 | 3653993 | 3654002 | 9 | |
| Deletion | Chr11 | 6096748 | 6096756 | 8 | |
| Deletion | Chr3 | 7245670 | 7245682 | 12 | |
| Deletion | Chr5 | 8398885 | 8398940 | 55 | |
| Deletion | Chr2 | 8718814 | 8718819 | 5 | |
| Deletion | Chr11 | 8961759 | 8961763 | 4 | |
| Deletion | Chr7 | 9108191 | 9108192 | 1 | |
| Deletion | Chr1 | 10474333 | 10474335 | 2 | |
| Deletion | Chr11 | 11093707 | 11093717 | 10 | |
| Deletion | Chr11 | 11279152 | 11279157 | 5 | |
| Deletion | Chr6 | 12373292 | 12373293 | 1 | |
| Deletion | Chr12 | 13162099 | 13162117 | 18 | |
| Deletion | Chr12 | 13389593 | 13397215 | 7622 | |
| Deletion | Chr5 | 14759368 | 14759369 | 1 | |
| Deletion | Chr10 | 15163225 | 15163240 | 15 | |
| Deletion | Chr10 | 18033409 | 18033419 | 10 | |
| Deletion | Chr10 | 19237647 | 19237655 | 8 | |
| Deletion | Chr9 | 21158586 | 21158595 | 9 | |
| Deletion | Chr11 | 21390195 | 21390205 | 10 | |
| Deletion | Chr8 | 21701378 | 21701380 | 2 | LOC_Os08g34550 |
| Deletion | Chr11 | 25530966 | 25530977 | 11 | |
| Deletion | Chr8 | 25943062 | 25943064 | 2 | |
| Deletion | Chr11 | 27608520 | 27608525 | 5 | LOC_Os11g45620 |
| Deletion | Chr8 | 27625588 | 27625590 | 2 | |
| Deletion | Chr2 | 31045486 | 31045487 | 1 | |
| Deletion | Chr3 | 34852434 | 34852437 | 3 | LOC_Os03g61410 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 18094826 | 18094826 | 1 | |
| Insertion | Chr10 | 19473448 | 19473449 | 2 | |
| Insertion | Chr11 | 16367368 | 16367369 | 2 | |
| Insertion | Chr6 | 4492630 | 4492632 | 3 | |
| Insertion | Chr7 | 22930902 | 22930903 | 2 | LOC_Os07g38240 |
| Insertion | Chr7 | 26267327 | 26267328 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 12300076 | 12478183 | |
| Inversion | Chr2 | 20193299 | 20259959 | |
| Inversion | Chr2 | 20193310 | 20260035 | |
| Inversion | Chr1 | 37155451 | 37345931 | LOC_Os01g63990 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 20260028 | Chr1 | 19348955 |
