Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1113-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1113-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 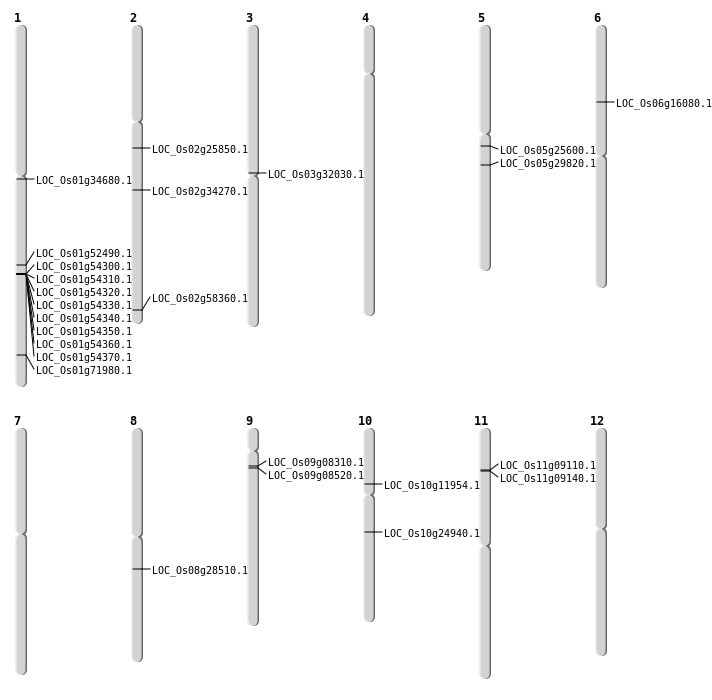
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15637375 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 19129183 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g34680 |
| SBS | Chr1 | 34231366 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 38507297 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 883213 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 11683368 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr10 | 13369523 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 6631492 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g11954 |
| SBS | Chr10 | 8106418 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 11059240 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 19669009 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 23527390 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 23527392 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 24093479 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 9294668 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 15145791 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g25850 |
| SBS | Chr2 | 15145793 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g25850 |
| SBS | Chr2 | 15160521 | C-T | HET | INTRON | |
| SBS | Chr2 | 27844461 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 9168336 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 14453042 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 16001464 | C-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 19410152 | A-G | HET | INTRON | |
| SBS | Chr3 | 30579571 | C-T | HET | INTRON | |
| SBS | Chr3 | 35426068 | C-T | HET | INTRON | |
| SBS | Chr3 | 5776600 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 5776605 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 5776613 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 14260246 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 14884147 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g25600 |
| SBS | Chr5 | 4325447 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 17591133 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 2277977 | C-T | HET | INTRON | |
| SBS | Chr6 | 29192966 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 3457816 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 3552740 | T-C | HET | INTRON | |
| SBS | Chr7 | 10850607 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 22984363 | C-G | HET | INTRON | |
| SBS | Chr8 | 1464617 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 17407965 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g28510 |
| SBS | Chr8 | 23965782 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 4444468 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g08520 |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 4296116 | 4296120 | 4 | LOC_Os09g08310 |
| Deletion | Chr2 | 4440576 | 4440596 | 20 | |
| Deletion | Chr11 | 4851207 | 4851226 | 19 | LOC_Os11g09110 |
| Deletion | Chr12 | 8215862 | 8215878 | 16 | |
| Deletion | Chr6 | 9158790 | 9158791 | 1 | LOC_Os06g16080 |
| Deletion | Chr10 | 12841166 | 12841174 | 8 | LOC_Os10g24940 |
| Deletion | Chr10 | 16801385 | 16801393 | 8 | |
| Deletion | Chr5 | 17260866 | 17260879 | 13 | LOC_Os05g29820 |
| Deletion | Chr7 | 18052680 | 18052683 | 3 | |
| Deletion | Chr3 | 18319183 | 18319187 | 4 | LOC_Os03g32030 |
| Deletion | Chr3 | 20102755 | 20102756 | 1 | |
| Deletion | Chr2 | 20499233 | 20499240 | 7 | LOC_Os02g34270 |
| Deletion | Chr7 | 22785965 | 22785977 | 12 | |
| Deletion | Chr6 | 29192238 | 29192241 | 3 | |
| Deletion | Chr1 | 30158160 | 30158190 | 30 | LOC_Os01g52490 |
| Deletion | Chr1 | 31254316 | 31284604 | 30288 | 8 |
| Deletion | Chr2 | 35695658 | 35695672 | 14 | LOC_Os02g58360 |
| Deletion | Chr1 | 41715994 | 41716012 | 18 | LOC_Os01g71980 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23239268 | 23239268 | 1 | |
| Insertion | Chr11 | 4866717 | 4866718 | 2 | LOC_Os11g09140 |
| Insertion | Chr3 | 21038827 | 21038827 | 1 | |
| Insertion | Chr4 | 28640503 | 28640507 | 5 | |
| Insertion | Chr5 | 25672977 | 25672978 | 2 | |
| Insertion | Chr7 | 17560179 | 17560180 | 2 | |
| Insertion | Chr7 | 8746903 | 8746906 | 4 | |
| Insertion | Chr9 | 9768729 | 9768734 | 6 |
No Inversion
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 8904593 | Chr10 | 18963097 |
