Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1114-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1114-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 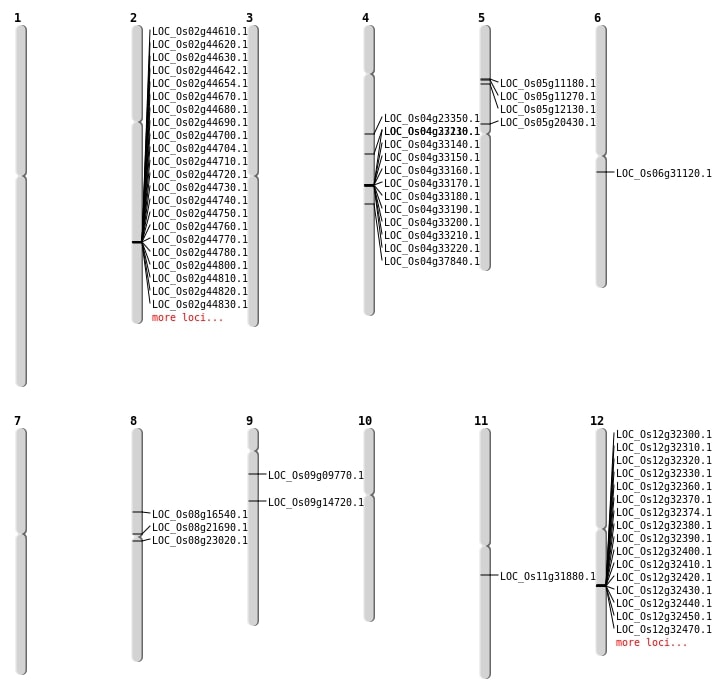
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14258265 | T-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 1535644 | T-A | HET | INTRON | |
| SBS | Chr1 | 29775991 | A-C | HET | UTR_3_PRIME | |
| SBS | Chr1 | 38676085 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 42071508 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 18194352 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 19388055 | A-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 11209195 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 16713140 | C-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 27361102 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 7596610 | C-T | HET | INTRON | |
| SBS | Chr11 | 9851443 | C-T | HOMO | INTRON | |
| SBS | Chr12 | 8263853 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 20212545 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 5489954 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 17795438 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 21472654 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 22001628 | T-C | HOMO | UTR_3_PRIME | |
| SBS | Chr3 | 30711627 | G-A | HOMO | UTR_5_PRIME | |
| SBS | Chr3 | 6901543 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 7452832 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr4 | 13334128 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g23350 |
| SBS | Chr4 | 2047128 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 22496349 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g37840 |
| SBS | Chr4 | 27034946 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 29993243 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 11988951 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g20430 |
| SBS | Chr5 | 17847683 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 29682585 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 9676319 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 15637748 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 17129451 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 2458484 | T-G | HET | UTR_3_PRIME | |
| SBS | Chr8 | 13863063 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g23020 |
| SBS | Chr8 | 20791448 | G-A | HET | INTRON | |
| SBS | Chr8 | 24938089 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 10778597 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 10928387 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 15765432 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 17453514 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 17615727 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 4621560 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 5292647 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g09770 |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 3460572 | 3460579 | 7 | |
| Deletion | Chr5 | 3687663 | 3688657 | 994 | |
| Deletion | Chr7 | 4218634 | 4218660 | 26 | |
| Deletion | Chr2 | 6053139 | 6053143 | 4 | |
| Deletion | Chr4 | 6682823 | 6682824 | 1 | |
| Deletion | Chr10 | 7659224 | 7659227 | 3 | |
| Deletion | Chr9 | 8742648 | 8742680 | 32 | LOC_Os09g14720 |
| Deletion | Chr2 | 9443849 | 9443850 | 1 | |
| Deletion | Chr4 | 10798595 | 10798596 | 1 | |
| Deletion | Chr8 | 12961307 | 12961314 | 7 | LOC_Os08g21690 |
| Deletion | Chr4 | 12961367 | 12961368 | 1 | |
| Deletion | Chr11 | 18729031 | 18729037 | 6 | LOC_Os11g31880 |
| Deletion | Chr12 | 19497001 | 20428000 | 931000 | 146 |
| Deletion | Chr12 | 19500573 | 19500574 | 1 | |
| Deletion | Chr4 | 20054001 | 20100000 | 46000 | 10 |
| Deletion | Chr5 | 20330422 | 20330424 | 2 | |
| Deletion | Chr2 | 27035001 | 27204000 | 169000 | 28 |
| Deletion | Chr1 | 31013160 | 31013161 | 1 | |
| Deletion | Chr1 | 38624596 | 38624598 | 2 |
Insertions: 10
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 6313859 | 7817976 | LOC_Os05g11180 |
| Inversion | Chr5 | 6313872 | 7817979 | LOC_Os05g11180 |
| Inversion | Chr5 | 6675120 | 6935828 | LOC_Os05g12130 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 6366368 | Chr4 | 16083640 | 2 |
| Translocation | Chr8 | 10125732 | Chr6 | 18090352 | 2 |
