Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1118-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1118-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 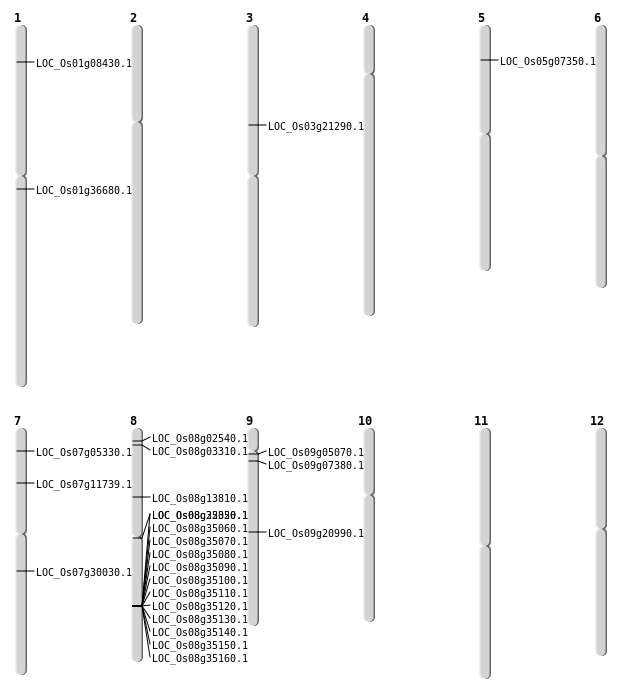
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21474460 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 40941249 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 4147612 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g08430 |
| SBS | Chr1 | 42750093 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 10691514 | G-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 11931771 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 26109145 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 26109147 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 26757239 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 10764920 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 21019966 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 23230333 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 24448807 | G-T | HET | INTRON | |
| SBS | Chr12 | 26501376 | A-T | HET | INTRON | |
| SBS | Chr2 | 10831772 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 21749871 | A-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 32319212 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 17091817 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 22128158 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 31106056 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 4588688 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 9416477 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 27679859 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 14188232 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 21472089 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 3917085 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g07350 |
| SBS | Chr5 | 501105 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 21151493 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 6394367 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 17699183 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g30030 |
| SBS | Chr7 | 23356973 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 2431066 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g05330 |
| SBS | Chr7 | 270904 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 8109862 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 13454298 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g22320 |
| SBS | Chr8 | 22065262 | C-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 8314448 | A-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 16040651 | G-T | HET | INTRON | |
| SBS | Chr9 | 16040652 | C-T | HET | INTRON | |
| SBS | Chr9 | 16040653 | C-T | HET | INTRON | |
| SBS | Chr9 | 2729982 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g05070 |
| SBS | Chr9 | 3673900 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07380 |
| SBS | Chr9 | 9507675 | G-A | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1049987 | 1050003 | 16 | LOC_Os08g02540 |
| Deletion | Chr12 | 1143230 | 1143231 | 1 | |
| Deletion | Chr8 | 1537356 | 1537364 | 8 | LOC_Os08g03310 |
| Deletion | Chr10 | 2497864 | 2497867 | 3 | |
| Deletion | Chr3 | 2510252 | 2510261 | 9 | |
| Deletion | Chr3 | 2696519 | 2696522 | 3 | |
| Deletion | Chr9 | 4014987 | 4015003 | 16 | |
| Deletion | Chr6 | 5751061 | 5751062 | 1 | |
| Deletion | Chr8 | 6099845 | 6099848 | 3 | |
| Deletion | Chr9 | 6186645 | 6186659 | 14 | |
| Deletion | Chr7 | 6495317 | 6495322 | 5 | LOC_Os07g11739 |
| Deletion | Chr12 | 6779157 | 6779159 | 2 | |
| Deletion | Chr3 | 12182055 | 12182070 | 15 | LOC_Os03g21290 |
| Deletion | Chr1 | 12320377 | 12320467 | 90 | |
| Deletion | Chr3 | 18233320 | 18233321 | 1 | |
| Deletion | Chr2 | 19577666 | 19577669 | 3 | |
| Deletion | Chr1 | 20359435 | 20359446 | 11 | |
| Deletion | Chr1 | 20365152 | 20371612 | 6460 | LOC_Os01g36680 |
| Deletion | Chr8 | 21764032 | 21764229 | 197 | |
| Deletion | Chr8 | 22037951 | 22037981 | 30 | |
| Deletion | Chr8 | 22093001 | 22163000 | 70000 | 12 |
| Deletion | Chr2 | 23454798 | 23454805 | 7 | |
| Deletion | Chr6 | 25744128 | 25744136 | 8 | |
| Deletion | Chr4 | 27022970 | 27022973 | 3 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 15380454 | 15380454 | 1 | |
| Insertion | Chr6 | 5911655 | 5911655 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 12648447 | 12902171 | LOC_Os09g20990 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 8247826 | Chr7 | 21480608 | LOC_Os08g13810 |
| Translocation | Chr8 | 8248645 | Chr7 | 21480604 | |
| Translocation | Chr11 | 11959380 | Chr5 | 8143225 |
