Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1136-19-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1136-19-S Alignment File |
| Seed Availability | No |
| Mapping (Hover to Zoom-In) | 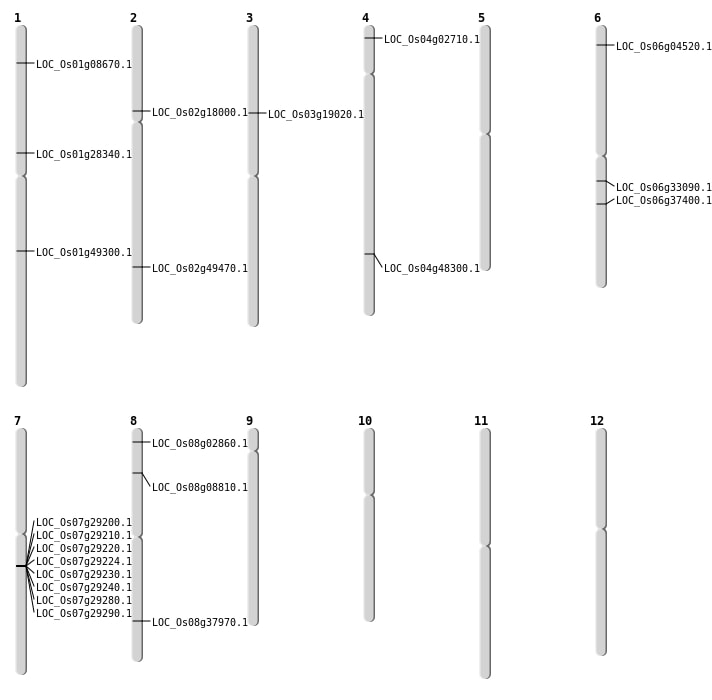
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 912205 | G-A | Homo | SYNONYMOUS_CODING | |
| SBS | Chr10 | 9560492 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 13926627 | G-A | Homo | INTERGENIC | |
| SBS | Chr11 | 19921311 | T-C | Homo | INTERGENIC | |
| SBS | Chr11 | 3053068 | G-A | Homo | INTERGENIC | |
| SBS | Chr12 | 10326775 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 24726212 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 7344007 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr12 | 9640610 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 15103372 | C-T | Homo | INTRON | |
| SBS | Chr2 | 24965518 | A-G | Homo | INTERGENIC | |
| SBS | Chr3 | 14210824 | T-A | Homo | INTERGENIC | |
| SBS | Chr3 | 17404739 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 30909504 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 11828566 | G-T | Homo | INTRON | |
| SBS | Chr4 | 17676636 | G-A | Homo | INTERGENIC | |
| SBS | Chr4 | 5642611 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 6740712 | C-G | Homo | INTERGENIC | |
| SBS | Chr5 | 17143876 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 3469450 | G-A | Homo | INTERGENIC | |
| SBS | Chr6 | 25744566 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 26931469 | G-C | Homo | INTERGENIC | |
| SBS | Chr6 | 29223883 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 14192400 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 2102163 | C-G | Homo | INTRON | |
| SBS | Chr7 | 4986564 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr8 | 10418243 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 24052391 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g37970 |
| SBS | Chr9 | 8522638 | G-A | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 154850 | 154851 | 1 | |
| Deletion | Chr4 | 1038789 | 1038790 | 1 | LOC_Os04g02710 |
| Deletion | Chr1 | 1279529 | 1279536 | 7 | |
| Deletion | Chr4 | 2216724 | 2216744 | 20 | |
| Deletion | Chr7 | 4927296 | 4927297 | 1 | |
| Deletion | Chr8 | 5107890 | 5107892 | 2 | LOC_Os08g08810 |
| Deletion | Chr11 | 5399302 | 5399312 | 10 | |
| Deletion | Chr5 | 6026519 | 6026528 | 9 | |
| Deletion | Chr3 | 6749526 | 6749529 | 3 | |
| Deletion | Chr8 | 7384220 | 7384222 | 2 | |
| Deletion | Chr2 | 10444822 | 10444828 | 6 | LOC_Os02g18000 |
| Deletion | Chr1 | 11032744 | 11032745 | 1 | |
| Deletion | Chr9 | 11794984 | 11794988 | 4 | |
| Deletion | Chr12 | 14657836 | 14657840 | 4 | |
| Deletion | Chr10 | 15928220 | 15928221 | 1 | |
| Deletion | Chr7 | 17095062 | 17095074 | 12 | |
| Deletion | Chr7 | 17097001 | 17163000 | 66000 | 8 |
| Deletion | Chr10 | 18444832 | 18444833 | 1 | |
| Deletion | Chr11 | 19072979 | 19072994 | 15 | |
| Deletion | Chr7 | 19087684 | 19087693 | 9 | |
| Deletion | Chr10 | 20593737 | 20593739 | 2 | |
| Deletion | Chr6 | 22109419 | 22109467 | 48 | LOC_Os06g37400 |
| Deletion | Chr12 | 22587247 | 22587248 | 1 | |
| Deletion | Chr4 | 25736705 | 25736711 | 6 | |
| Deletion | Chr4 | 25942367 | 25942368 | 1 | |
| Deletion | Chr5 | 26867597 | 26867598 | 1 | |
| Deletion | Chr2 | 27244815 | 27244820 | 5 | |
| Deletion | Chr4 | 28787244 | 28787253 | 9 | LOC_Os04g48300 |
| Deletion | Chr2 | 33674464 | 33674466 | 2 | |
| Deletion | Chr1 | 41342107 | 41342108 | 1 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11341825 | 11341867 | 43 | |
| Insertion | Chr1 | 15877751 | 15877751 | 1 | LOC_Os01g28340 |
| Insertion | Chr1 | 4317984 | 4317984 | 1 | LOC_Os01g08670 |
| Insertion | Chr10 | 5013984 | 5013985 | 2 | |
| Insertion | Chr2 | 31487063 | 31487064 | 2 | |
| Insertion | Chr3 | 477662 | 477662 | 1 | |
| Insertion | Chr3 | 7548789 | 7548830 | 42 | |
| Insertion | Chr4 | 16253951 | 16253956 | 6 | |
| Insertion | Chr5 | 6390419 | 6390419 | 1 | |
| Insertion | Chr6 | 5682267 | 5682269 | 3 | |
| Insertion | Chr8 | 15143567 | 15143568 | 2 | |
| Insertion | Chr9 | 15342636 | 15342636 | 1 |
No Inversion
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 1208324 | Chr3 | 10652855 | 2 |
| Translocation | Chr8 | 1209411 | Chr3 | 10652850 | 2 |
| Translocation | Chr12 | 1644038 | Chr6 | 19264972 | LOC_Os06g33090 |
| Translocation | Chr6 | 1948397 | Chr1 | 28341754 | 2 |
| Translocation | Chr6 | 1948792 | Chr1 | 28341778 | LOC_Os01g49300 |
| Translocation | Chr12 | 4365544 | Chr2 | 30232469 | LOC_Os02g49470 |
| Translocation | Chr7 | 5606803 | Chr2 | 18079972 | |
| Translocation | Chr11 | 7272741 | Chr7 | 17129582 | LOC_Os07g29224 |
| Translocation | Chr9 | 10934296 | Chr5 | 936258 |
