Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1158-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1158-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 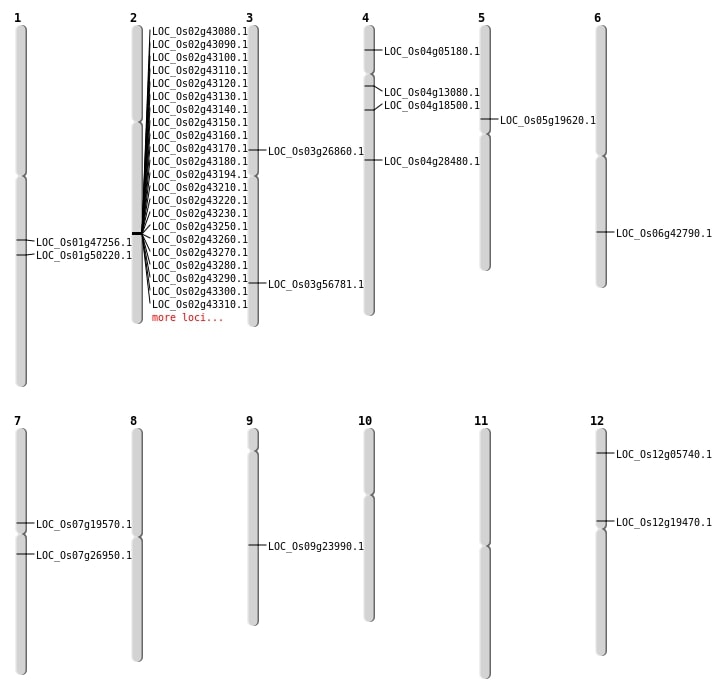
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22966002 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 25534455 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 28855081 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g50220 |
| SBS | Chr1 | 41173373 | G-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 17868090 | C-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr11 | 16254741 | T-C | HOMO | INTRON | |
| SBS | Chr11 | 778173 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 10910550 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 27296795 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 30635764 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 3998001 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 22196267 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 10227603 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18500 |
| SBS | Chr4 | 7214724 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g13080 |
| SBS | Chr4 | 902975 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 11434443 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g19620 |
| SBS | Chr5 | 11715263 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 12122345 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 20577511 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 7611148 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 7629709 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 20251235 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 514819 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 11598160 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g19570 |
| SBS | Chr7 | 15864705 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 161006 | A-T | HET | INTRON | |
| SBS | Chr8 | 14574924 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 2642328 | T-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 4076991 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 5709653 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 9914107 | G-A | HET | INTRON |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 2647858 | 2647859 | 1 | LOC_Os12g05740 |
| Deletion | Chr4 | 6229608 | 6229609 | 1 | |
| Deletion | Chr7 | 6490020 | 6490021 | 1 | |
| Deletion | Chr4 | 6619016 | 6619020 | 4 | |
| Deletion | Chr7 | 8388856 | 8388858 | 2 | |
| Deletion | Chr4 | 9872204 | 9872206 | 2 | |
| Deletion | Chr12 | 11321044 | 11321061 | 17 | LOC_Os12g19470 |
| Deletion | Chr6 | 13355997 | 13356017 | 20 | |
| Deletion | Chr11 | 14208264 | 14208265 | 1 | |
| Deletion | Chr3 | 14506974 | 14506975 | 1 | |
| Deletion | Chr8 | 15513385 | 15513386 | 1 | |
| Deletion | Chr7 | 15611388 | 15611390 | 2 | LOC_Os07g26950 |
| Deletion | Chr4 | 16869480 | 16869492 | 12 | LOC_Os04g28480 |
| Deletion | Chr6 | 16876149 | 16876159 | 10 | |
| Deletion | Chr10 | 18070036 | 18070037 | 1 | |
| Deletion | Chr2 | 19060162 | 19060171 | 9 | |
| Deletion | Chr8 | 19231246 | 19231247 | 1 | |
| Deletion | Chr4 | 19785111 | 19787195 | 2084 | |
| Deletion | Chr4 | 20507885 | 20507886 | 1 | |
| Deletion | Chr3 | 20834848 | 20834852 | 4 | |
| Deletion | Chr1 | 22456557 | 22456563 | 6 | |
| Deletion | Chr3 | 23075213 | 23075229 | 16 | |
| Deletion | Chr2 | 24244397 | 24249487 | 5090 | |
| Deletion | Chr5 | 24430252 | 24430253 | 1 | |
| Deletion | Chr11 | 24450373 | 24450379 | 6 | |
| Deletion | Chr2 | 25943001 | 26150000 | 207000 | 26 |
| Deletion | Chr3 | 32348219 | 32348225 | 6 | LOC_Os03g56781 |
| Deletion | Chr4 | 34642142 | 34642158 | 16 |
Insertions: 10
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 25189673 | 27007488 | LOC_Os01g47256 |
| Inversion | Chr1 | 25534418 | 27007488 | LOC_Os01g47256 |
| Inversion | Chr6 | 25722165 | 25726874 | LOC_Os06g42790 |
| Inversion | Chr6 | 25722181 | 25734754 | LOC_Os06g42790 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 9584239 | Chr2 | 24248787 | |
| Translocation | Chr3 | 9584243 | Chr2 | 24244545 | |
| Translocation | Chr5 | 11445668 | Chr2 | 26026995 | |
| Translocation | Chr5 | 11445695 | Chr2 | 26004102 | |
| Translocation | Chr9 | 14290346 | Chr4 | 2584783 | 2 |
| Translocation | Chr3 | 15322616 | Chr1 | 39128731 | LOC_Os03g26860 |
| Translocation | Chr11 | 21327652 | Chr3 | 25354092 |
