Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1163-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1163-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 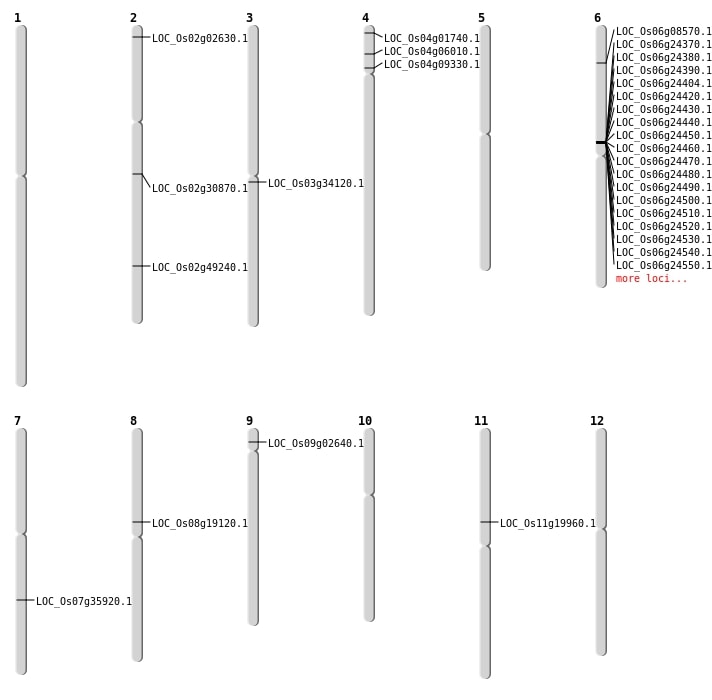
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11870165 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 1226725 | C-T | HET | INTRON | |
| SBS | Chr1 | 31842365 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 21380667 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 21566349 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 11490094 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g19960 |
| SBS | Chr11 | 24647912 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 5622240 | C-T | HET | INTRON | |
| SBS | Chr11 | 9681300 | C-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 11468824 | T-G | HOMO | UTR_3_PRIME | |
| SBS | Chr12 | 14891193 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 25342887 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 4795600 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 29536200 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 5104092 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 7882285 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 8308357 | T-C | HET | INTRON | |
| SBS | Chr2 | 965950 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g02630 |
| SBS | Chr3 | 19440863 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g34120 |
| SBS | Chr4 | 26353955 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 3116779 | C-T | HET | STOP_GAINED | LOC_Os04g06010 |
| SBS | Chr4 | 484907 | C-T | HET | STOP_GAINED | LOC_Os04g01740 |
| SBS | Chr4 | 5917251 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 18752289 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 23383618 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 15453432 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 16905057 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 26789212 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g44380 |
| SBS | Chr7 | 15188118 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 21909550 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 22800297 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 9666345 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 11430356 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g19120 |
| SBS | Chr8 | 14451542 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 22511862 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 23667308 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 8775559 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 12540451 | T-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 7038180 | G-A | HET | INTERGENIC |
Deletions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1146826 | 1146827 | 1 | LOC_Os09g02640 |
| Deletion | Chr7 | 2421712 | 2421725 | 13 | |
| Deletion | Chr6 | 4268144 | 4268154 | 10 | LOC_Os06g08570 |
| Deletion | Chr5 | 5345564 | 5345565 | 1 | |
| Deletion | Chr2 | 5476532 | 5476542 | 10 | |
| Deletion | Chr9 | 6577115 | 6577116 | 1 | |
| Deletion | Chr8 | 9001504 | 9001557 | 53 | |
| Deletion | Chr9 | 12900897 | 12900912 | 15 | |
| Deletion | Chr6 | 14269092 | 15234106 | 965014 | 155 |
| Deletion | Chr1 | 14716283 | 14716284 | 1 | |
| Deletion | Chr9 | 16116159 | 16116161 | 2 | |
| Deletion | Chr5 | 22032624 | 22032625 | 1 | |
| Deletion | Chr4 | 29350594 | 29350597 | 3 | |
| Deletion | Chr2 | 30099779 | 30099784 | 5 | LOC_Os02g49240 |
No Insertion
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 18419787 | 19148923 | LOC_Os02g30870 |
| Inversion | Chr7 | 21495121 | 21753678 | LOC_Os07g35920 |
| Inversion | Chr6 | 25260942 | 25377932 | LOC_Os06g42050 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 4958334 | Chr2 | 19148926 | LOC_Os04g09330 |
| Translocation | Chr4 | 4958373 | Chr2 | 19173238 | LOC_Os04g09330 |
