Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1175-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1175-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 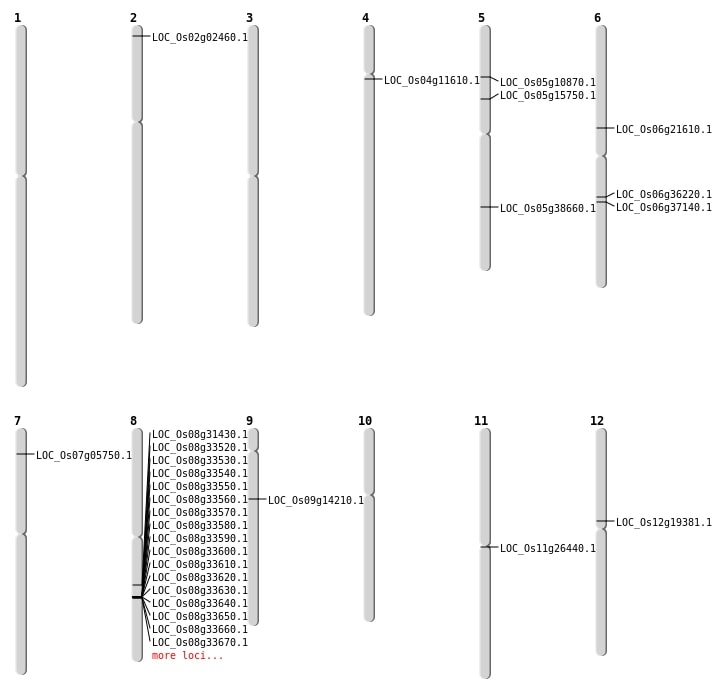
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24764052 | T-A | HOMO | INTRON | |
| SBS | Chr1 | 28205897 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 5684221 | G-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 15270847 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 16607758 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 5226465 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 15015015 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 15141761 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g26440 |
| SBS | Chr11 | 16716145 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 8642139 | A-T | HET | INTRON | |
| SBS | Chr12 | 11037531 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 13682840 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 10482799 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 21493248 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 867636 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g02460 |
| SBS | Chr3 | 34352986 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 14317018 | A-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 31530372 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 32813990 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 34438609 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr4 | 6355387 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g11610 |
| SBS | Chr5 | 10128886 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 6022541 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g10870 |
| SBS | Chr5 | 6022542 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 12469864 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g21610 |
| SBS | Chr6 | 16539010 | G-A | HET | INTRON | |
| SBS | Chr6 | 16743810 | A-T | HET | INTRON | |
| SBS | Chr6 | 21216214 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g36220 |
| SBS | Chr6 | 29863262 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 2767723 | C-T | HET | STOP_GAINED | LOC_Os07g05750 |
| SBS | Chr7 | 9080541 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 19441610 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g31430 |
| SBS | Chr8 | 24058962 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 7073146 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 12234193 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 13722277 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 13805119 | C-G | HET | INTRON | |
| SBS | Chr9 | 3019723 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 8398667 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g14210 |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 413707 | 413801 | 94 | |
| Deletion | Chr5 | 3390398 | 3390400 | 2 | |
| Deletion | Chr4 | 3517231 | 3517241 | 10 | |
| Deletion | Chr1 | 4995990 | 4995995 | 5 | |
| Deletion | Chr5 | 8891288 | 8891292 | 4 | LOC_Os05g15750 |
| Deletion | Chr8 | 9704549 | 9704550 | 1 | |
| Deletion | Chr8 | 13126709 | 13126743 | 34 | |
| Deletion | Chr5 | 16748196 | 16748208 | 12 | |
| Deletion | Chr5 | 18763230 | 18763249 | 19 | |
| Deletion | Chr5 | 20014329 | 20014330 | 1 | |
| Deletion | Chr10 | 20441475 | 20441499 | 24 | |
| Deletion | Chr8 | 20919857 | 21038835 | 118978 | 18 |
| Deletion | Chr10 | 21168309 | 21168311 | 2 | |
| Deletion | Chr6 | 21940120 | 21944140 | 4020 | LOC_Os06g37140 |
| Deletion | Chr3 | 22468987 | 22468996 | 9 | |
| Deletion | Chr5 | 22665839 | 22665841 | 2 | LOC_Os05g38660 |
| Deletion | Chr8 | 23016836 | 23016837 | 1 | |
| Deletion | Chr10 | 23160169 | 23160186 | 17 | |
| Deletion | Chr4 | 25168718 | 25168723 | 5 | |
| Deletion | Chr8 | 26120024 | 26120025 | 1 | |
| Deletion | Chr7 | 26461964 | 26461966 | 2 | |
| Deletion | Chr8 | 27595578 | 27595581 | 3 | |
| Deletion | Chr1 | 35076329 | 35076330 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 23712853 | 23712853 | 1 | |
| Insertion | Chr3 | 3502138 | 3502138 | 1 | |
| Insertion | Chr7 | 11427287 | 11427288 | 2 | |
| Insertion | Chr9 | 14985480 | 14985481 | 2 |
No Inversion
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 11272892 | Chr7 | 2280247 | LOC_Os12g19381 |
