Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1180-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1180-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 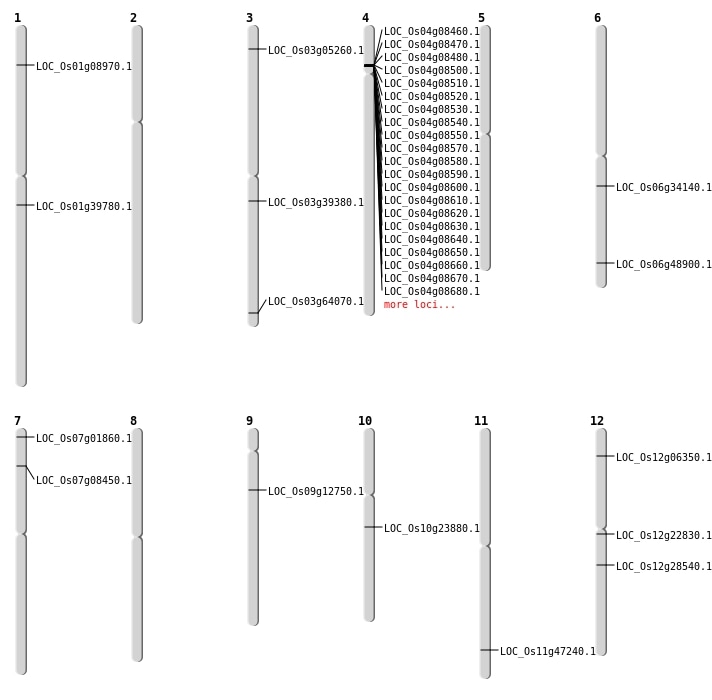
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14067499 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 14067500 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 31771745 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 4501178 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g08970 |
| SBS | Chr1 | 7883468 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 14142662 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 14721465 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 14721468 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 14721469 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 18257987 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 4420206 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 607856 | A-G | HET | INTRON | |
| SBS | Chr11 | 24265271 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 28410559 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g47240 |
| SBS | Chr12 | 11337847 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 17470454 | T-C | HET | INTRON | |
| SBS | Chr12 | 22740839 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 22740840 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 27462709 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 22627144 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr2 | 24064181 | G-T | HET | INTRON | |
| SBS | Chr2 | 8758003 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 12171817 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 19390804 | A-T | HET | INTRON | |
| SBS | Chr3 | 36204771 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g64070 |
| SBS | Chr3 | 4642220 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 11454773 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 124212 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 1293402 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 29605416 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g48900 |
| SBS | Chr7 | 23995247 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 18534959 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 20465571 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 14633705 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 3643453 | A-G | HET | INTERGENIC |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 318229 | 318242 | 13 | |
| Deletion | Chr7 | 517779 | 517803 | 24 | LOC_Os07g01860 |
| Deletion | Chr3 | 1688880 | 1688883 | 3 | |
| Deletion | Chr9 | 2680558 | 2680559 | 1 | |
| Deletion | Chr12 | 3046827 | 3046833 | 6 | LOC_Os12g06350 |
| Deletion | Chr12 | 3732594 | 3732596 | 2 | |
| Deletion | Chr7 | 4353636 | 4353659 | 23 | LOC_Os07g08450 |
| Deletion | Chr4 | 4553001 | 4765000 | 212000 | 29 |
| Deletion | Chr2 | 7267610 | 7267614 | 4 | |
| Deletion | Chr4 | 7934058 | 7934074 | 16 | |
| Deletion | Chr12 | 8959330 | 8959697 | 367 | |
| Deletion | Chr4 | 10597102 | 10597117 | 15 | LOC_Os04g19090 |
| Deletion | Chr10 | 11715561 | 11715562 | 1 | |
| Deletion | Chr12 | 12887319 | 12887337 | 18 | LOC_Os12g22830 |
| Deletion | Chr10 | 12895971 | 12895979 | 8 | |
| Deletion | Chr12 | 14073700 | 14073707 | 7 | |
| Deletion | Chr11 | 15059509 | 15059522 | 13 | |
| Deletion | Chr7 | 15297024 | 15297027 | 3 | |
| Deletion | Chr7 | 15766509 | 15766535 | 26 | |
| Deletion | Chr11 | 16715984 | 16715985 | 1 | |
| Deletion | Chr12 | 16869013 | 16869023 | 10 | LOC_Os12g28540 |
| Deletion | Chr10 | 17846975 | 17847058 | 83 | |
| Deletion | Chr12 | 18063065 | 18063069 | 4 | |
| Deletion | Chr4 | 20775209 | 20775210 | 1 | |
| Deletion | Chr3 | 21884408 | 21884941 | 533 | LOC_Os03g39380 |
| Deletion | Chr12 | 21893907 | 21893913 | 6 | |
| Deletion | Chr1 | 22438351 | 22438369 | 18 | LOC_Os01g39780 |
| Deletion | Chr4 | 23495939 | 23495947 | 8 | |
| Deletion | Chr5 | 24552067 | 24552070 | 3 | |
| Deletion | Chr11 | 25048840 | 25048852 | 12 | |
| Deletion | Chr4 | 27020967 | 27020971 | 4 | |
| Deletion | Chr1 | 29443764 | 29443765 | 1 | |
| Deletion | Chr1 | 31967686 | 31967687 | 1 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 7829346 | 7829347 | 2 | |
| Insertion | Chr6 | 22672325 | 22672327 | 3 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 2546228 | 3227659 | LOC_Os03g05260 |
| Inversion | Chr4 | 7086952 | 7779855 | LOC_Os04g12830 |
| Inversion | Chr10 | 7376853 | 7404646 | |
| Inversion | Chr10 | 12232295 | 12250137 | LOC_Os10g23880 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 60358 | Chr7 | 19204207 | |
| Translocation | Chr9 | 7322267 | Chr4 | 7090501 | LOC_Os09g12750 |
| Translocation | Chr9 | 7322285 | Chr4 | 7086978 | 2 |
| Translocation | Chr6 | 19887580 | Chr4 | 8795475 | 2 |
