Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1182-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1182-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 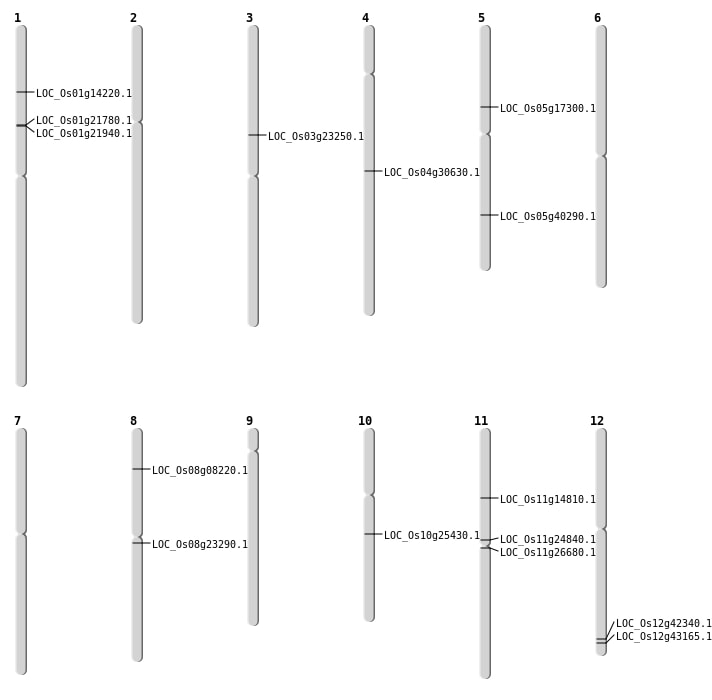
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16265525 | G-A | HET | INTRON | |
| SBS | Chr1 | 22798621 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 26407381 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 9713005 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 9701916 | C-T | HET | INTRON | |
| SBS | Chr11 | 14164428 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g24840 |
| SBS | Chr11 | 14851168 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 11022263 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 26325271 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g42340 |
| SBS | Chr12 | 26806715 | A-G | HET | START_GAINED | LOC_Os12g43165 |
| SBS | Chr12 | 4630026 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 9298985 | G-A | HET | INTRON | |
| SBS | Chr2 | 14737621 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 14743144 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 14743146 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 14593751 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 19924289 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 21563873 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 31495606 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 34355514 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 34479253 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 7332449 | A-G | HET | INTRON | |
| SBS | Chr4 | 14223349 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 293100 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 33511067 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 22997167 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 3637393 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 3292372 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 14057839 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g23290 |
| SBS | Chr8 | 17331515 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 27374355 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 28083442 | T-G | HET | UTR_3_PRIME | |
| SBS | Chr8 | 4704050 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g08220 |
| SBS | Chr8 | 4704051 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g08220 |
| SBS | Chr8 | 801625 | G-T | HET | INTERGENIC |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 912337 | 912352 | 15 | |
| Deletion | Chr6 | 1453901 | 1453907 | 6 | |
| Deletion | Chr3 | 2792144 | 2792151 | 7 | |
| Deletion | Chr5 | 4294256 | 4294257 | 1 | |
| Deletion | Chr11 | 6004952 | 6004961 | 9 | |
| Deletion | Chr3 | 6186854 | 6187044 | 190 | |
| Deletion | Chr4 | 6534202 | 6534205 | 3 | |
| Deletion | Chr9 | 6555189 | 6555194 | 5 | |
| Deletion | Chr3 | 7851413 | 7851425 | 12 | |
| Deletion | Chr11 | 8334248 | 8334279 | 31 | LOC_Os11g14810 |
| Deletion | Chr9 | 9164291 | 9164294 | 3 | |
| Deletion | Chr5 | 9210066 | 9210076 | 10 | |
| Deletion | Chr5 | 9911791 | 9911792 | 1 | LOC_Os05g17300 |
| Deletion | Chr1 | 10327296 | 10327300 | 4 | |
| Deletion | Chr9 | 11067871 | 11067879 | 8 | |
| Deletion | Chr10 | 11209388 | 11209389 | 1 | |
| Deletion | Chr1 | 12120046 | 12120055 | 9 | |
| Deletion | Chr10 | 13150079 | 13151894 | 1815 | LOC_Os10g25430 |
| Deletion | Chr11 | 15283788 | 15283809 | 21 | LOC_Os11g26680 |
| Deletion | Chr11 | 15321034 | 15321050 | 16 | |
| Deletion | Chr10 | 15530878 | 15530879 | 1 | |
| Deletion | Chr10 | 17329253 | 17329255 | 2 | |
| Deletion | Chr3 | 17489188 | 17489190 | 2 | |
| Deletion | Chr11 | 18182890 | 18182905 | 15 | |
| Deletion | Chr4 | 18294655 | 18294661 | 6 | LOC_Os04g30630 |
| Deletion | Chr5 | 21247874 | 21247882 | 8 | |
| Deletion | Chr5 | 21504463 | 21504468 | 5 | |
| Deletion | Chr3 | 21518639 | 21518641 | 2 | |
| Deletion | Chr2 | 22134810 | 22134811 | 1 | |
| Deletion | Chr3 | 23209191 | 23209192 | 1 | |
| Deletion | Chr5 | 23673720 | 23673722 | 2 | LOC_Os05g40290 |
| Deletion | Chr1 | 25511755 | 25511760 | 5 | |
| Deletion | Chr2 | 31053226 | 31053242 | 16 | |
| Deletion | Chr4 | 34606894 | 34606908 | 14 |
Insertions: 8
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 7973626 | 8087350 | LOC_Os01g14220 |
| Inversion | Chr1 | 7973638 | 7976641 | LOC_Os01g14220 |
| Inversion | Chr1 | 12221176 | 12304548 | 2 |
| Inversion | Chr1 | 12221187 | 12304551 | 2 |
| Inversion | Chr3 | 13458080 | 13459178 | LOC_Os03g23250 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 3234375 | Chr1 | 23346976 | |
| Translocation | Chr5 | 6994875 | Chr3 | 6186858 | |
| Translocation | Chr12 | 9390181 | Chr10 | 13150088 | |
| Translocation | Chr12 | 9390182 | Chr10 | 13151935 | LOC_Os10g25430 |
