Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1195-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1195-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 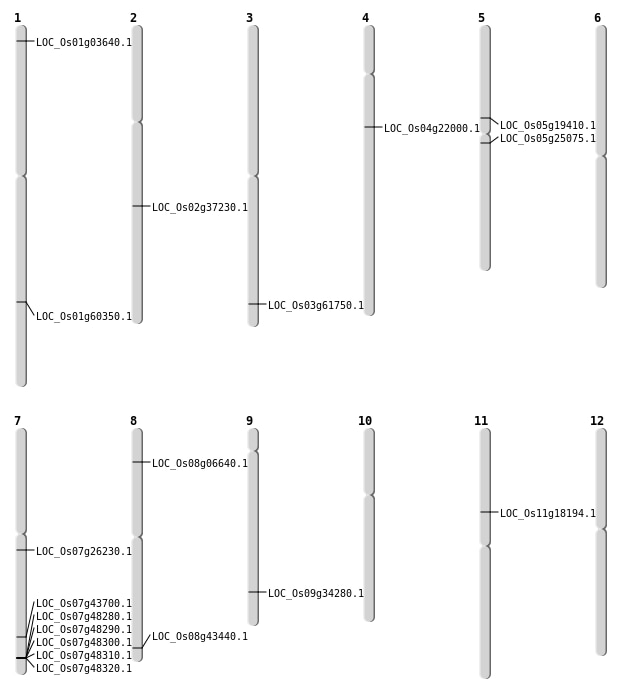
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 25857218 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 17238789 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 8390090 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 8408472 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 9589519 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 16938955 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 21558925 | C-T | HET | INTRON | |
| SBS | Chr12 | 343085 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 15391046 | C-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 22496060 | C-T | HET | STOP_GAINED | LOC_Os02g37230 |
| SBS | Chr2 | 9357543 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 9398748 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 24340384 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 35005040 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g61750 |
| SBS | Chr4 | 10878421 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 20099815 | C-A | HET | UTR_3_PRIME | |
| SBS | Chr4 | 22171790 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 27620660 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 32079662 | C-A | HET | INTRON | |
| SBS | Chr4 | 4466505 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 8059781 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 11273821 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g19410 |
| SBS | Chr5 | 14519585 | G-A | HOMO | STOP_GAINED | LOC_Os05g25075 |
| SBS | Chr5 | 19440501 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 22591086 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 4749572 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 15291845 | C-A | HET | INTRON | |
| SBS | Chr6 | 21250340 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 27083294 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 14458177 | A-G | HET | INTRON | |
| SBS | Chr7 | 27634002 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 27634009 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 28769454 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 11862572 | C-T | HOMO | INTERGENIC |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 77056 | 77071 | 15 | |
| Deletion | Chr7 | 906755 | 906757 | 2 | |
| Deletion | Chr8 | 1387124 | 1387133 | 9 | |
| Deletion | Chr1 | 1500162 | 1500167 | 5 | LOC_Os01g03640 |
| Deletion | Chr12 | 2008815 | 2008816 | 1 | |
| Deletion | Chr8 | 3744297 | 3744305 | 8 | LOC_Os08g06640 |
| Deletion | Chr6 | 5030961 | 5030962 | 1 | |
| Deletion | Chr7 | 5072529 | 5072539 | 10 | |
| Deletion | Chr2 | 9397425 | 9397429 | 4 | |
| Deletion | Chr11 | 10258847 | 10258856 | 9 | LOC_Os11g18194 |
| Deletion | Chr1 | 11453312 | 11453315 | 3 | |
| Deletion | Chr1 | 12384718 | 12384726 | 8 | |
| Deletion | Chr4 | 12456617 | 12456623 | 6 | LOC_Os04g22000 |
| Deletion | Chr7 | 14472456 | 14472459 | 3 | |
| Deletion | Chr7 | 15090901 | 15090909 | 8 | LOC_Os07g26230 |
| Deletion | Chr5 | 15651126 | 15651128 | 2 | |
| Deletion | Chr12 | 16287401 | 16287411 | 10 | |
| Deletion | Chr7 | 16637466 | 16637472 | 6 | |
| Deletion | Chr7 | 22866074 | 22866083 | 9 | |
| Deletion | Chr3 | 23021974 | 23021979 | 5 | |
| Deletion | Chr8 | 24747735 | 24747743 | 8 | |
| Deletion | Chr12 | 24919715 | 24919729 | 14 | |
| Deletion | Chr8 | 25030865 | 25030878 | 13 | |
| Deletion | Chr7 | 26155505 | 26155910 | 405 | LOC_Os07g43700 |
| Deletion | Chr8 | 27479812 | 27479822 | 10 | LOC_Os08g43440 |
| Deletion | Chr7 | 27514313 | 27514324 | 11 | |
| Deletion | Chr2 | 27808932 | 27808943 | 11 | |
| Deletion | Chr2 | 27833212 | 27833213 | 1 | |
| Deletion | Chr4 | 27944235 | 27944239 | 4 | |
| Deletion | Chr7 | 28570149 | 28570156 | 7 | |
| Deletion | Chr7 | 28840001 | 28878000 | 38000 | 5 |
| Deletion | Chr3 | 32099332 | 32099333 | 1 | |
| Deletion | Chr2 | 33355568 | 33355569 | 1 | |
| Deletion | Chr1 | 34910844 | 34910845 | 1 | LOC_Os01g60350 |
Insertions: 8
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 2440444 | 2980854 | |
| Inversion | Chr8 | 24530378 | 24585668 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 14223729 | Chr10 | 19141714 | |
| Translocation | Chr9 | 20238904 | Chr7 | 28877537 | 2 |
| Translocation | Chr9 | 20238913 | Chr7 | 28839877 | LOC_Os09g34280 |
