Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1200-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1200-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 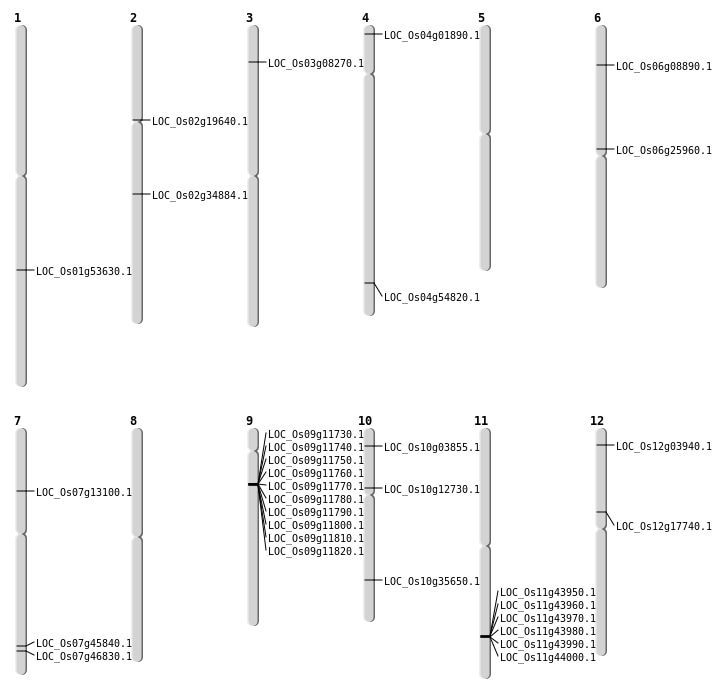
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16821881 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 28205993 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 28681884 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 28683683 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 11660991 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1752726 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g03855 |
| SBS | Chr10 | 7101634 | C-T | HET | STOP_GAINED | LOC_Os10g12730 |
| SBS | Chr10 | 8461801 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 20711333 | G-A | HET | INTRON | |
| SBS | Chr11 | 24994204 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 10158061 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g17740 |
| SBS | Chr12 | 11102992 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 7299139 | T-A | HET | INTRON | |
| SBS | Chr2 | 11483564 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g19640 |
| SBS | Chr2 | 17185826 | G-T | HET | INTRON | |
| SBS | Chr2 | 2314944 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 34064845 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 9866120 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 10090292 | C-A | HOMO | UTR_3_PRIME | |
| SBS | Chr3 | 11911917 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 11911918 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 15779830 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 4430011 | T-C | HET | INTRON | |
| SBS | Chr4 | 24750525 | G-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 561836 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g01890 |
| SBS | Chr5 | 9966649 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 15199099 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g25960 |
| SBS | Chr6 | 4474661 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g08890 |
| SBS | Chr7 | 12472711 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 24148303 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 27358506 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g45840 |
| SBS | Chr7 | 7502150 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g13100 |
| SBS | Chr8 | 10207433 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 5465877 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 7524896 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 18353547 | G-A | HET | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 37048 | 37055 | 7 | |
| Deletion | Chr8 | 1474157 | 1474178 | 21 | |
| Deletion | Chr6 | 3611908 | 3611916 | 8 | |
| Deletion | Chr3 | 4204168 | 4204171 | 3 | LOC_Os03g08270 |
| Deletion | Chr9 | 6547001 | 6628000 | 81000 | 10 |
| Deletion | Chr8 | 7743055 | 7743056 | 1 | |
| Deletion | Chr10 | 9005111 | 9005121 | 10 | |
| Deletion | Chr3 | 9539577 | 9539688 | 111 | |
| Deletion | Chr6 | 10134044 | 10134045 | 1 | |
| Deletion | Chr11 | 13620120 | 13622103 | 1983 | |
| Deletion | Chr8 | 13756319 | 13758181 | 1862 | |
| Deletion | Chr10 | 17210951 | 17210952 | 1 | |
| Deletion | Chr8 | 18532540 | 18532563 | 23 | |
| Deletion | Chr10 | 19067801 | 19067816 | 15 | LOC_Os10g35650 |
| Deletion | Chr10 | 19341003 | 19341011 | 8 | |
| Deletion | Chr6 | 20320987 | 20320997 | 10 | |
| Deletion | Chr2 | 20921507 | 20921508 | 1 | LOC_Os02g34884 |
| Deletion | Chr1 | 21744740 | 21744751 | 11 | |
| Deletion | Chr9 | 22592251 | 22592252 | 1 | |
| Deletion | Chr8 | 23146456 | 23146457 | 1 | |
| Deletion | Chr7 | 24032023 | 24032031 | 8 | |
| Deletion | Chr4 | 24425027 | 24425053 | 26 | |
| Deletion | Chr11 | 26549001 | 26584000 | 35000 | 6 |
| Deletion | Chr1 | 28289332 | 28289338 | 6 | |
| Deletion | Chr7 | 29072714 | 29072719 | 5 | |
| Deletion | Chr1 | 30791862 | 30791867 | 5 | LOC_Os01g53630 |
| Deletion | Chr3 | 33701971 | 33701972 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 2611697 | 2611700 | 4 | |
| Insertion | Chr3 | 181913 | 181914 | 2 | |
| Insertion | Chr8 | 10173562 | 10173563 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 11520916 | 12003241 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 1631027 | Chr2 | 30991348 | LOC_Os12g03940 |
| Translocation | Chr9 | 6546359 | Chr4 | 32602088 | 2 |
| Translocation | Chr9 | 6628231 | Chr4 | 32602362 | LOC_Os04g54820 |
| Translocation | Chr11 | 9031546 | Chr7 | 23098246 | |
| Translocation | Chr7 | 27991710 | Chr4 | 32602326 | 2 |
| Translocation | Chr7 | 27991726 | Chr4 | 33052298 | LOC_Os07g46830 |
