Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1212-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1212-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 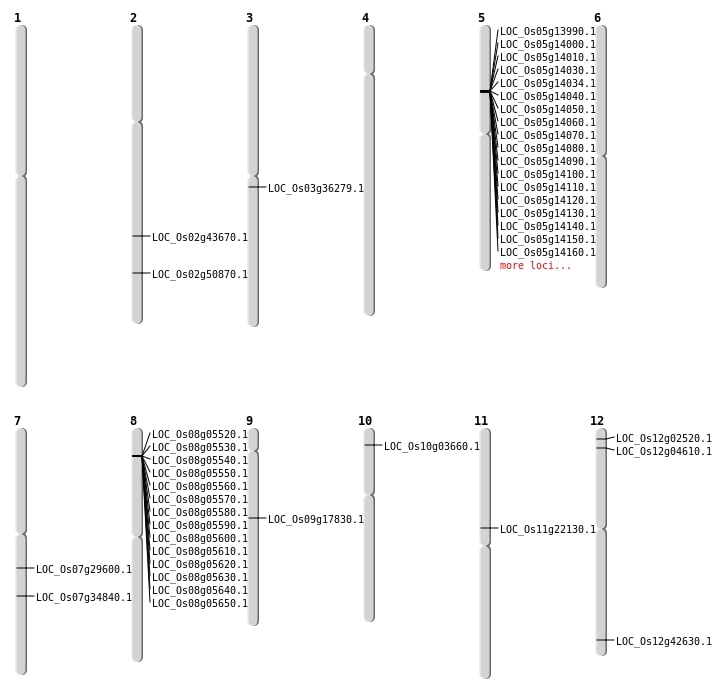
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21079068 | C-A | HOMO | INTRON | |
| SBS | Chr1 | 23430748 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 2520175 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 30089231 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 5684422 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 3057724 | A-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 7391556 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 1535574 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 17569240 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 26498362 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g42630 |
| SBS | Chr12 | 3082450 | G-A | HET | INTRON | |
| SBS | Chr2 | 3891138 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 1951916 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 20125116 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g36279 |
| SBS | Chr3 | 33122697 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 7171370 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 10136687 | C-A | HOMO | INTRON | |
| SBS | Chr4 | 2348137 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 4122763 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 1915288 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 19473398 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 21255140 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 9344531 | T-G | HET | INTERGENIC | |
| SBS | Chr6 | 10077526 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 17447440 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 29159055 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 5830808 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 17399975 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr7 | 17399976 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr7 | 17399977 | C-T | HET | START_GAINED | LOC_Os07g29600 |
| SBS | Chr8 | 13740200 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr8 | 20574369 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 20574370 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 22457172 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 25357486 | T-C | HET | INTRON | |
| SBS | Chr8 | 3645113 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 13901185 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 21419153 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 4206769 | C-T | HET | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 163652 | 163659 | 7 | |
| Deletion | Chr12 | 771835 | 771839 | 4 | |
| Deletion | Chr12 | 859324 | 859334 | 10 | LOC_Os12g02520 |
| Deletion | Chr12 | 1094964 | 1094973 | 9 | |
| Deletion | Chr10 | 1632346 | 1632348 | 2 | LOC_Os10g03660 |
| Deletion | Chr5 | 1915283 | 1915284 | 1 | |
| Deletion | Chr12 | 1949714 | 1949715 | 1 | LOC_Os12g04610 |
| Deletion | Chr8 | 2942341 | 3018345 | 76004 | 14 |
| Deletion | Chr6 | 4466133 | 4466134 | 1 | |
| Deletion | Chr5 | 7788001 | 8022000 | 234000 | 29 |
| Deletion | Chr8 | 10819410 | 10819414 | 4 | |
| Deletion | Chr5 | 12099918 | 12099924 | 6 | LOC_Os05g20620 |
| Deletion | Chr8 | 14344382 | 14344391 | 9 | |
| Deletion | Chr10 | 14628250 | 14628253 | 3 | |
| Deletion | Chr2 | 17644490 | 17644495 | 5 | |
| Deletion | Chr5 | 17829070 | 17829071 | 1 | |
| Deletion | Chr10 | 18899571 | 18899572 | 1 | |
| Deletion | Chr7 | 19026030 | 19026032 | 2 | |
| Deletion | Chr11 | 19245687 | 19245718 | 31 | |
| Deletion | Chr5 | 20372001 | 20430000 | 58000 | 7 |
| Deletion | Chr7 | 20881824 | 20881825 | 1 | LOC_Os07g34840 |
| Deletion | Chr2 | 26353002 | 26353011 | 9 | LOC_Os02g43670 |
| Deletion | Chr6 | 28982661 | 28982662 | 1 | |
| Deletion | Chr2 | 31089299 | 31089311 | 12 | LOC_Os02g50870 |
| Deletion | Chr4 | 31409347 | 31409353 | 6 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 17811477 | 17811478 | 2 | |
| Insertion | Chr1 | 41964925 | 41964925 | 1 | |
| Insertion | Chr3 | 26816364 | 26816370 | 7 | |
| Insertion | Chr8 | 15238874 | 15238875 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 6641008 | 6655885 | |
| Inversion | Chr4 | 9861855 | 10427402 | |
| Inversion | Chr9 | 10161664 | 10908042 | LOC_Os09g17830 |
| Inversion | Chr5 | 23104037 | 24564975 | LOC_Os05g39380 |
| Inversion | Chr5 | 28262324 | 28402034 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 7922203 | Chr4 | 9861884 | |
| Translocation | Chr11 | 12699557 | Chr7 | 14859313 | LOC_Os11g22130 |
| Translocation | Chr5 | 20431787 | Chr4 | 10427339 |
