Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1217-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1217-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 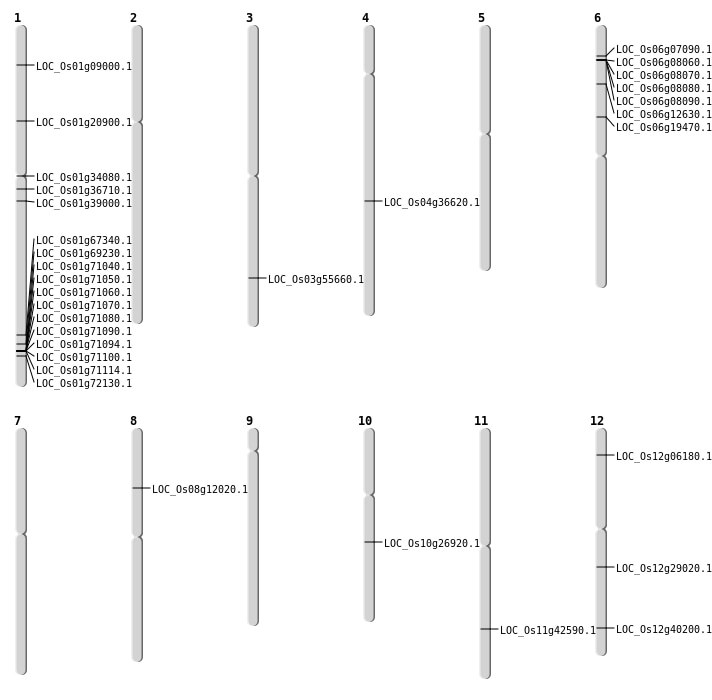
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11678409 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g20900 |
| SBS | Chr1 | 14302883 | C-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 16279541 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 31053666 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 39092256 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g67340 |
| SBS | Chr1 | 4518844 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g09000 |
| SBS | Chr1 | 4637693 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 14158806 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g26920 |
| SBS | Chr10 | 14158807 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g26920 |
| SBS | Chr11 | 15828703 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 16466299 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 18849139 | C-A | HET | INTRON | |
| SBS | Chr11 | 18876977 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr11 | 21421620 | C-T | HOMO | INTRON | |
| SBS | Chr11 | 22477490 | T-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 8258990 | C-G | HET | INTRON | |
| SBS | Chr12 | 13322949 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 1548998 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 17174697 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g29020 |
| SBS | Chr12 | 17824523 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 21203347 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 21558308 | G-A | HET | INTRON | |
| SBS | Chr12 | 22938089 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 10489766 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 26291702 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 34293079 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 34675256 | T-C | HOMO | INTRON | |
| SBS | Chr2 | 4760785 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 7764911 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 8967717 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 14477892 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 1453812 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 10481035 | T-G | HET | INTERGENIC | |
| SBS | Chr5 | 15435342 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 314898 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 1706646 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 3902442 | C-T | HET | INTRON | |
| SBS | Chr7 | 6543619 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 8201247 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 13095815 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 16705853 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 3129017 | T-A | HOMO | UTR_3_PRIME | |
| SBS | Chr8 | 7057034 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g12020 |
| SBS | Chr9 | 9490704 | G-A | HET | INTRON | |
| SBS | Chr9 | 9545857 | A-T | HET | SYNONYMOUS_CODING |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 1213522 | 1213523 | 1 | |
| Deletion | Chr1 | 3748487 | 3748492 | 5 | |
| Deletion | Chr6 | 3902273 | 3925254 | 22981 | 4 |
| Deletion | Chr10 | 4057622 | 4057623 | 1 | |
| Deletion | Chr6 | 5947338 | 5947347 | 9 | |
| Deletion | Chr5 | 6830152 | 6830154 | 2 | |
| Deletion | Chr9 | 9475252 | 9475438 | 186 | |
| Deletion | Chr4 | 9873214 | 9873217 | 3 | |
| Deletion | Chr4 | 11725992 | 11726011 | 19 | |
| Deletion | Chr7 | 11854117 | 11854120 | 3 | |
| Deletion | Chr11 | 15849242 | 15849243 | 1 | |
| Deletion | Chr10 | 16418067 | 16418089 | 22 | |
| Deletion | Chr12 | 17617841 | 17617852 | 11 | |
| Deletion | Chr5 | 17967772 | 17967786 | 14 | |
| Deletion | Chr8 | 18354488 | 18354548 | 60 | |
| Deletion | Chr2 | 18465574 | 18465579 | 5 | |
| Deletion | Chr1 | 18763504 | 18763507 | 3 | LOC_Os01g34080 |
| Deletion | Chr9 | 19231524 | 19231529 | 5 | |
| Deletion | Chr11 | 19625427 | 19625436 | 9 | |
| Deletion | Chr11 | 20007733 | 20007742 | 9 | |
| Deletion | Chr7 | 20955688 | 20955689 | 1 | |
| Deletion | Chr5 | 21254511 | 21254517 | 6 | |
| Deletion | Chr3 | 21801110 | 21801118 | 8 | |
| Deletion | Chr7 | 22026632 | 22026640 | 8 | |
| Deletion | Chr9 | 22311274 | 22311279 | 5 | |
| Deletion | Chr11 | 24225150 | 24225169 | 19 | |
| Deletion | Chr12 | 24263056 | 24263059 | 3 | |
| Deletion | Chr11 | 24675781 | 24681717 | 5936 | |
| Deletion | Chr6 | 27504568 | 27504569 | 1 | |
| Deletion | Chr6 | 27768279 | 27768291 | 12 | |
| Deletion | Chr3 | 29872873 | 29872951 | 78 | |
| Deletion | Chr1 | 31232996 | 31233000 | 4 | |
| Deletion | Chr3 | 31691233 | 31691246 | 13 | LOC_Os03g55660 |
| Deletion | Chr1 | 31720975 | 31720977 | 2 | |
| Deletion | Chr1 | 41112001 | 41165000 | 53000 | 10 |
Insertions: 6
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 3385020 | 11090620 | 2 |
| Inversion | Chr6 | 3705675 | 11090625 | LOC_Os06g19470 |
| Inversion | Chr1 | 20383214 | 21927041 | 2 |
| Inversion | Chr1 | 20383233 | 21927043 | 2 |
| Inversion | Chr1 | 40238164 | 41842156 | 2 |
| Inversion | Chr1 | 41111644 | 41842146 | LOC_Os01g72130 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2930092 | Chr6 | 6890642 | 2 |
| Translocation | Chr12 | 2930677 | Chr6 | 6890643 | 2 |
| Translocation | Chr12 | 22729739 | Chr1 | 24726876 | |
| Translocation | Chr11 | 24681710 | Chr4 | 22088431 | LOC_Os04g36620 |
| Translocation | Chr11 | 24681711 | Chr4 | 22088432 | LOC_Os04g36620 |
| Translocation | Chr12 | 24883650 | Chr11 | 6038590 | LOC_Os12g40200 |
| Translocation | Chr8 | 25532905 | Chr6 | 3925293 | LOC_Os06g08090 |
| Translocation | Chr8 | 25532924 | Chr6 | 3902278 | LOC_Os06g08060 |
| Translocation | Chr11 | 25635009 | Chr9 | 9475262 | LOC_Os11g42590 |
