Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1234-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1234-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 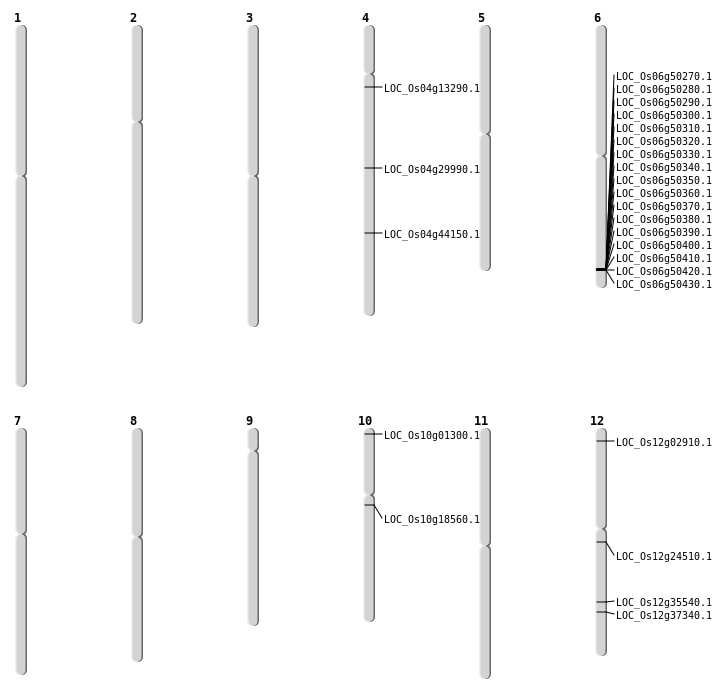
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17707742 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 24008688 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 24274675 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 24274677 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 24274682 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 7116169 | A-T | HOMO | INTRON | |
| SBS | Chr10 | 14137163 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 192464 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g01300 |
| SBS | Chr11 | 3584676 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 21612120 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g35540 |
| SBS | Chr12 | 22929920 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g37340 |
| SBS | Chr12 | 8290436 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 26133989 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 5039916 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 2759424 | A-C | HET | INTRON | |
| SBS | Chr3 | 2873936 | T-G | HOMO | INTRON | |
| SBS | Chr3 | 35791502 | G-T | HET | INTRON | |
| SBS | Chr4 | 26134272 | G-A | HET | START_GAINED | LOC_Os04g44150 |
| SBS | Chr4 | 7489504 | C-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 21075115 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 23159277 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 28795040 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 477531 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 7091693 | G-T | HET | INTRON | |
| SBS | Chr6 | 10602735 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 20480431 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 25514018 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 737847 | A-G | HET | INTRON | |
| SBS | Chr7 | 8154077 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 18463189 | A-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 2712917 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 8689628 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 8689629 | G-T | HET | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 994222 | 994230 | 8 | |
| Deletion | Chr12 | 1075210 | 1075219 | 9 | LOC_Os12g02910 |
| Deletion | Chr8 | 3411872 | 3411876 | 4 | |
| Deletion | Chr9 | 5389928 | 5389929 | 1 | |
| Deletion | Chr4 | 7371559 | 7371560 | 1 | LOC_Os04g13290 |
| Deletion | Chr12 | 7421772 | 7421773 | 1 | |
| Deletion | Chr1 | 7508275 | 7508276 | 1 | |
| Deletion | Chr12 | 13991746 | 13991748 | 2 | LOC_Os12g24510 |
| Deletion | Chr7 | 16229695 | 16229698 | 3 | |
| Deletion | Chr8 | 19141774 | 19141784 | 10 | |
| Deletion | Chr5 | 19627434 | 19627447 | 13 | |
| Deletion | Chr8 | 21061055 | 21061060 | 5 | |
| Deletion | Chr10 | 22151296 | 22151301 | 5 | |
| Deletion | Chr5 | 23217713 | 23217714 | 1 | |
| Deletion | Chr6 | 24177563 | 24177565 | 2 | |
| Deletion | Chr1 | 24652235 | 24652236 | 1 | |
| Deletion | Chr11 | 26963584 | 26963585 | 1 | |
| Deletion | Chr6 | 30432001 | 30530000 | 98000 | 17 |
| Deletion | Chr3 | 33657943 | 33657944 | 1 | |
| Deletion | Chr1 | 38372119 | 38372143 | 24 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 7861130 | 7861130 | 1 | |
| Insertion | Chr10 | 9413777 | 9413778 | 2 | LOC_Os10g18560 |
| Insertion | Chr11 | 4357002 | 4357003 | 2 | |
| Insertion | Chr12 | 4576485 | 4576485 | 1 | |
| Insertion | Chr2 | 16516655 | 16516656 | 2 |
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 5149211 | Chr1 | 1991889 | |
| Translocation | Chr9 | 20883363 | Chr4 | 17888294 | LOC_Os04g29990 |
