Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1255-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1255-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 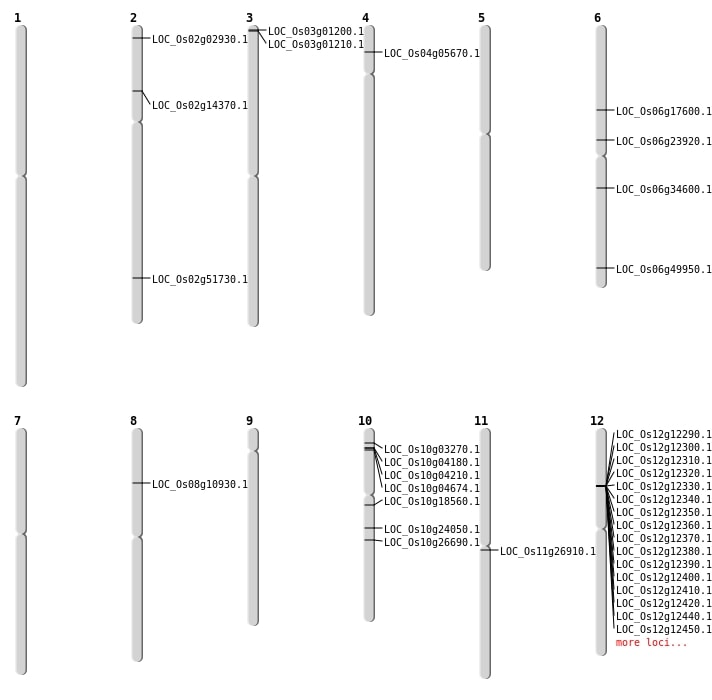
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 2833367 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 22344923 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 4661040 | G-A | HET | INTRON | |
| SBS | Chr11 | 8307460 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 11213577 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 24562825 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 3178830 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 10657964 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 15614316 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 18630266 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 18831211 | A-C | HET | INTRON | |
| SBS | Chr2 | 19517941 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 20894544 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 27743059 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 23762089 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 16075082 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 2150088 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 2900462 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g05670 |
| SBS | Chr4 | 3398560 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 5707434 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 17393812 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 19861246 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 5948143 | G-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 13976482 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13976483 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g23920 |
| SBS | Chr6 | 16155059 | G-T | HOMO | INTRON | |
| SBS | Chr6 | 20121910 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g34600 |
| SBS | Chr6 | 30245568 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g49950 |
| SBS | Chr6 | 5215382 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 10475650 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 16812434 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 26760017 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 1618445 | A-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 1060712 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 2005047 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 7306124 | G-A | HET | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 126346 | 135892 | 9546 | 2 |
| Deletion | Chr2 | 1149134 | 1149137 | 3 | LOC_Os02g02930 |
| Deletion | Chr10 | 1385516 | 1385518 | 2 | LOC_Os10g03270 |
| Deletion | Chr12 | 1509324 | 1509332 | 8 | |
| Deletion | Chr8 | 1709754 | 1709765 | 11 | |
| Deletion | Chr10 | 1952001 | 1965000 | 13000 | 2 |
| Deletion | Chr5 | 3211630 | 3211634 | 4 | |
| Deletion | Chr6 | 3345667 | 3345668 | 1 | |
| Deletion | Chr2 | 5287242 | 5287244 | 2 | |
| Deletion | Chr10 | 5503738 | 5503740 | 2 | |
| Deletion | Chr12 | 5751543 | 5751545 | 2 | |
| Deletion | Chr12 | 6752001 | 6873000 | 121000 | 18 |
| Deletion | Chr12 | 6875001 | 7099000 | 224000 | 37 |
| Deletion | Chr3 | 6922736 | 6922739 | 3 | |
| Deletion | Chr2 | 7884171 | 7884178 | 7 | LOC_Os02g14370 |
| Deletion | Chr3 | 8079604 | 8079622 | 18 | |
| Deletion | Chr3 | 8120088 | 8120096 | 8 | |
| Deletion | Chr3 | 11796670 | 11796696 | 26 | |
| Deletion | Chr10 | 12321727 | 12321733 | 6 | LOC_Os10g24050 |
| Deletion | Chr3 | 14811415 | 14811424 | 9 | |
| Deletion | Chr7 | 18517139 | 18517140 | 1 | |
| Deletion | Chr2 | 18629967 | 18629972 | 5 | |
| Deletion | Chr7 | 21421119 | 21421125 | 6 | |
| Deletion | Chr7 | 24198201 | 24198202 | 1 | |
| Deletion | Chr6 | 27411297 | 27411369 | 72 | |
| Deletion | Chr6 | 28015923 | 28015926 | 3 | |
| Deletion | Chr2 | 31687841 | 31687852 | 11 | LOC_Os02g51730 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 13939435 | 13939440 | 6 | LOC_Os10g26690 |
| Insertion | Chr10 | 9413777 | 9413778 | 2 | LOC_Os10g18560 |
| Insertion | Chr2 | 11595555 | 11595558 | 4 | |
| Insertion | Chr5 | 21765747 | 21765750 | 4 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 1952093 | 2228204 | 2 |
| Inversion | Chr8 | 6443611 | 20240543 | LOC_Os08g10930 |
| Inversion | Chr12 | 6751686 | 8121293 | |
| Inversion | Chr12 | 7099108 | 8121305 | LOC_Os12g12850 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 1965658 | Chr1 | 21680627 | |
| Translocation | Chr10 | 2228237 | Chr1 | 21681043 | LOC_Os10g04674 |
| Translocation | Chr9 | 11597067 | Chr6 | 10213537 | LOC_Os06g17600 |
| Translocation | Chr11 | 15462243 | Chr8 | 20240547 | LOC_Os11g26910 |
