Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1276-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1276-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 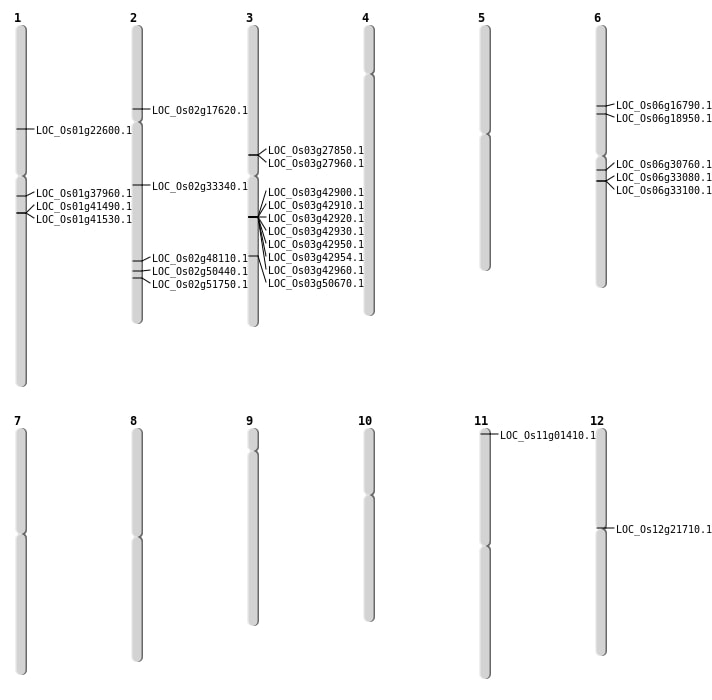
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12513187 | G-A | HET | INTRON | |
| SBS | Chr1 | 12705443 | C-T | HET | STOP_GAINED | LOC_Os01g22600 |
| SBS | Chr1 | 12705446 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g22600 |
| SBS | Chr1 | 21428496 | C-G | HOMO | INTRON | |
| SBS | Chr1 | 23486725 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g41490 |
| SBS | Chr1 | 23510580 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g41530 |
| SBS | Chr1 | 24656549 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 35725006 | C-A | HET | INTRON | |
| SBS | Chr1 | 8550017 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 10714027 | T-C | HOMO | INTRON | |
| SBS | Chr11 | 251587 | C-T | HET | STOP_GAINED | LOC_Os11g01410 |
| SBS | Chr11 | 9932550 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 14313511 | T-A | HOMO | INTRON | |
| SBS | Chr12 | 14519964 | G-C | HET | INTRON | |
| SBS | Chr12 | 1834684 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 29456864 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g48110 |
| SBS | Chr2 | 29560367 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 15999807 | G-A | HET | STOP_GAINED | LOC_Os03g27850 |
| SBS | Chr3 | 33141109 | C-A | HET | UTR_5_PRIME | |
| SBS | Chr3 | 7374020 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 17323104 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 19765440 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 2372976 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 24724607 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 29613226 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 8455549 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 17842721 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g30760 |
| SBS | Chr6 | 17842722 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 21091914 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 6031996 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 7797539 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 11939168 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 24043735 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 25773116 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 27864362 | A-T | HET | INTRON | |
| SBS | Chr8 | 7762677 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 13290748 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 20687753 | A-T | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 3568978 | 3568979 | 1 | |
| Deletion | Chr12 | 4053818 | 4053827 | 9 | |
| Deletion | Chr3 | 6568923 | 6568927 | 4 | |
| Deletion | Chr3 | 9780748 | 9780780 | 32 | |
| Deletion | Chr6 | 10771182 | 10777510 | 6328 | LOC_Os06g18950 |
| Deletion | Chr9 | 11402324 | 11402334 | 10 | |
| Deletion | Chr3 | 14047849 | 14047861 | 12 | |
| Deletion | Chr6 | 14755393 | 14755405 | 12 | |
| Deletion | Chr3 | 16064837 | 16064839 | 2 | LOC_Os03g27960 |
| Deletion | Chr7 | 18362807 | 18362809 | 2 | |
| Deletion | Chr1 | 18909171 | 18909177 | 6 | |
| Deletion | Chr2 | 19810593 | 19810601 | 8 | LOC_Os02g33340 |
| Deletion | Chr1 | 21266164 | 21266182 | 18 | LOC_Os01g37960 |
| Deletion | Chr5 | 22255775 | 22255776 | 1 | |
| Deletion | Chr4 | 22375135 | 22375136 | 1 | |
| Deletion | Chr1 | 22629754 | 22629773 | 19 | |
| Deletion | Chr8 | 22723214 | 22723225 | 11 | |
| Deletion | Chr3 | 23920001 | 23962000 | 42000 | 7 |
| Deletion | Chr3 | 28935341 | 28935348 | 7 | LOC_Os03g50670 |
| Deletion | Chr2 | 31698286 | 31698404 | 118 | LOC_Os02g51750 |
| Deletion | Chr3 | 33577469 | 33577488 | 19 | |
| Deletion | Chr2 | 35197756 | 35197770 | 14 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 25761349 | 25761349 | 1 | |
| Insertion | Chr2 | 10135304 | 10135307 | 4 | LOC_Os02g17620 |
| Insertion | Chr3 | 23671316 | 23671319 | 4 | |
| Insertion | Chr6 | 25246934 | 25246935 | 2 | |
| Insertion | Chr7 | 3165265 | 3165266 | 2 | |
| Insertion | Chr9 | 16276374 | 16276374 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 19259222 | 20004899 | LOC_Os06g33080 |
| Inversion | Chr6 | 19272359 | 20004896 | LOC_Os06g33100 |
| Inversion | Chr6 | 19272370 | 20004899 | LOC_Os06g33100 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3746094 | Chr1 | 12697758 | |
| Translocation | Chr11 | 3746096 | Chr1 | 12697553 | |
| Translocation | Chr6 | 5136746 | Chr2 | 30794803 | LOC_Os02g50440 |
| Translocation | Chr12 | 12207510 | Chr6 | 9724859 | 2 |
| Translocation | Chr12 | 12210644 | Chr6 | 9724843 | LOC_Os06g16790 |
| Translocation | Chr12 | 20981305 | Chr6 | 23038900 |
