Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1277-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1277-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 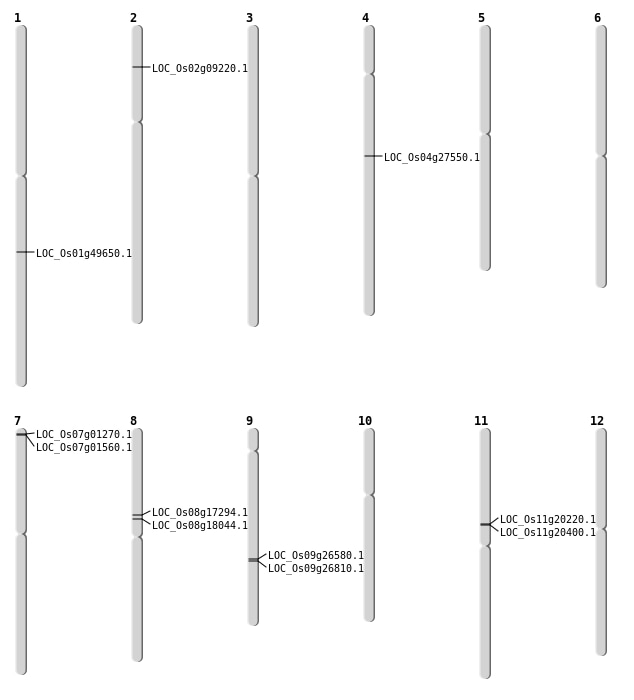
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11227977 | C-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 15873160 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 16860325 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 16860480 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 17727329 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 20215310 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 4574637 | A-T | HOMO | INTRON | |
| SBS | Chr11 | 11679980 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g20220 |
| SBS | Chr11 | 11811713 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g20400 |
| SBS | Chr11 | 16034828 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 16680939 | C-T | HOMO | INTRON | |
| SBS | Chr11 | 23742277 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 23742278 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 28878790 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 6159091 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 2302478 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 26628226 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 12139425 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 1416212 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 30611626 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 7037951 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 28148469 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 34044115 | T-C | HOMO | INTRON | |
| SBS | Chr4 | 10897137 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 35165654 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 26904105 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 28862907 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 29473392 | T-A | HET | INTRON | |
| SBS | Chr6 | 1259826 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 29717888 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 13331404 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 13331405 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 5877771 | T-C | HOMO | INTRON | |
| SBS | Chr8 | 10572877 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g17294 |
| SBS | Chr8 | 10572878 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g17294 |
| SBS | Chr8 | 13416753 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 16029746 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 16029748 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 20082779 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 9350135 | G-A | HET | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 448060 | 448068 | 8 | |
| Deletion | Chr10 | 773401 | 773405 | 4 | |
| Deletion | Chr6 | 2624204 | 2624205 | 1 | |
| Deletion | Chr11 | 2915583 | 2915589 | 6 | |
| Deletion | Chr2 | 3174515 | 3174519 | 4 | |
| Deletion | Chr2 | 4742892 | 4742893 | 1 | LOC_Os02g09220 |
| Deletion | Chr4 | 10887874 | 10887876 | 2 | |
| Deletion | Chr8 | 11060906 | 11060907 | 1 | LOC_Os08g18044 |
| Deletion | Chr10 | 13227481 | 13227491 | 10 | |
| Deletion | Chr5 | 14874259 | 14874260 | 1 | |
| Deletion | Chr12 | 15014898 | 15014902 | 4 | |
| Deletion | Chr4 | 16018442 | 16018448 | 6 | |
| Deletion | Chr4 | 16300169 | 16306188 | 6019 | LOC_Os04g27550 |
| Deletion | Chr11 | 19315697 | 19315703 | 6 | |
| Deletion | Chr8 | 20829488 | 20829492 | 4 | |
| Deletion | Chr10 | 21841362 | 21841656 | 294 | |
| Deletion | Chr5 | 22209900 | 22209906 | 6 | |
| Deletion | Chr9 | 22455036 | 22455048 | 12 | |
| Deletion | Chr10 | 22647953 | 22647958 | 5 | |
| Deletion | Chr9 | 22879024 | 22879036 | 12 | |
| Deletion | Chr5 | 22889017 | 22889019 | 2 | |
| Deletion | Chr3 | 23209265 | 23209266 | 1 | |
| Deletion | Chr8 | 27820285 | 27820287 | 2 | |
| Deletion | Chr1 | 28539420 | 28539421 | 1 | LOC_Os01g49650 |
| Deletion | Chr4 | 31489970 | 31490028 | 58 | |
| Deletion | Chr1 | 36522376 | 36522378 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 10308547 | 10308550 | 4 | |
| Insertion | Chr6 | 5048099 | 5048100 | 2 | |
| Insertion | Chr7 | 29661649 | 29661650 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 170133 | 352900 | 2 |
| Inversion | Chr9 | 16100825 | 16288196 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 9228980 | Chr4 | 7080219 | |
| Translocation | Chr10 | 21841637 | Chr3 | 3457171 |
