Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1319-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1319-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 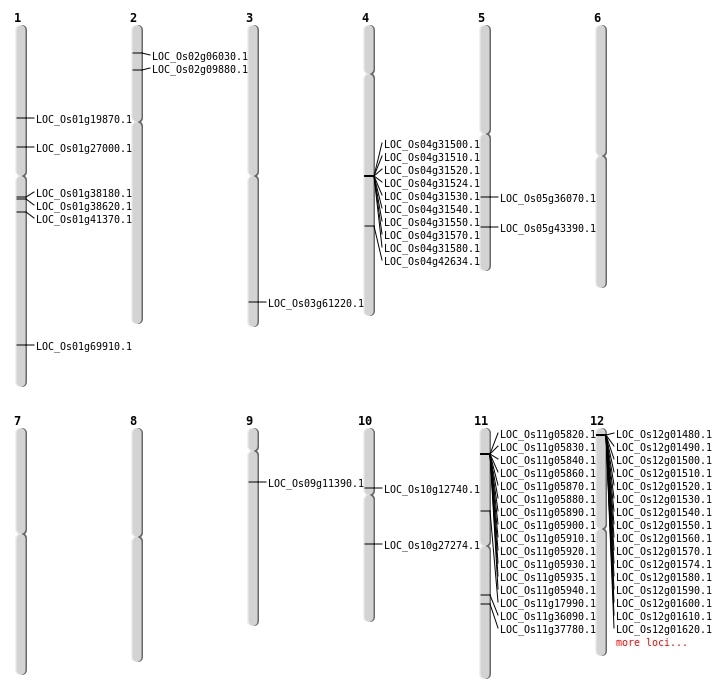
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11275533 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g19870 |
| SBS | Chr1 | 13081962 | T-A | HET | INTRON | |
| SBS | Chr1 | 15043653 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g27000 |
| SBS | Chr1 | 15319021 | G-T | HET | INTRON | |
| SBS | Chr1 | 22788183 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 31924378 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 40398194 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g69910 |
| SBS | Chr10 | 11106087 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 7107949 | G-A | HOMO | STOP_GAINED | LOC_Os10g12740 |
| SBS | Chr10 | 8420119 | G-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 10112687 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g17990 |
| SBS | Chr11 | 13656580 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 21295283 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 7247933 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 3200681 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 4231670 | G-A | HET | INTRON | |
| SBS | Chr2 | 26436624 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 32437785 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 32437789 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 34768910 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g61220 |
| SBS | Chr3 | 35725724 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 19633397 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 8251318 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 9757040 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 11379172 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 20553757 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 20553758 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 21376227 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g36070 |
| SBS | Chr5 | 25231421 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g43390 |
| SBS | Chr6 | 25469519 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 16009024 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 7486370 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 12786171 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 18184056 | C-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 26633750 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 1545168 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 6338280 | T-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr9 | 6338281 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr9 | 6338283 | C-A | HET | SPLICE_SITE_DONOR | LOC_Os09g11390 |
| SBS | Chr9 | 6971537 | T-C | HET | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 290001 | 1002000 | 712000 | 131 |
| Deletion | Chr11 | 903033 | 903035 | 2 | |
| Deletion | Chr6 | 1889168 | 1889173 | 5 | |
| Deletion | Chr11 | 2691001 | 2806000 | 115000 | 14 |
| Deletion | Chr4 | 2953421 | 2953424 | 3 | |
| Deletion | Chr2 | 3007457 | 3007459 | 2 | LOC_Os02g06030 |
| Deletion | Chr2 | 5106911 | 5113220 | 6309 | LOC_Os02g09880 |
| Deletion | Chr12 | 10844774 | 10844779 | 5 | |
| Deletion | Chr9 | 13662638 | 13662646 | 8 | |
| Deletion | Chr10 | 14397712 | 14397724 | 12 | LOC_Os10g27274 |
| Deletion | Chr9 | 14855360 | 14855364 | 4 | |
| Deletion | Chr5 | 17054920 | 17054923 | 3 | |
| Deletion | Chr5 | 18092888 | 18092892 | 4 | |
| Deletion | Chr2 | 18737684 | 18737690 | 6 | |
| Deletion | Chr4 | 18834001 | 18886000 | 52000 | 9 |
| Deletion | Chr12 | 19586423 | 19588759 | 2336 | |
| Deletion | Chr4 | 20483317 | 20483318 | 1 | |
| Deletion | Chr11 | 21215788 | 21215793 | 5 | LOC_Os11g36090 |
| Deletion | Chr4 | 21667816 | 21667817 | 1 | |
| Deletion | Chr11 | 22369871 | 22369882 | 11 | LOC_Os11g37780 |
| Deletion | Chr1 | 23410803 | 23410805 | 2 | LOC_Os01g41370 |
| Deletion | Chr1 | 24520259 | 24520260 | 1 | |
| Deletion | Chr4 | 25220498 | 25232153 | 11655 | LOC_Os04g42634 |
| Deletion | Chr12 | 25531957 | 25531967 | 10 | |
| Deletion | Chr7 | 25983398 | 25983399 | 1 | |
| Deletion | Chr1 | 31894666 | 31894680 | 14 | |
| Deletion | Chr4 | 33257619 | 33257620 | 1 | |
| Deletion | Chr3 | 33837386 | 33837389 | 3 |
Insertions: 6
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 21388748 | 21699414 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 25222941 | Chr1 | 21699418 | LOC_Os01g38620 |
| Translocation | Chr4 | 25229713 | Chr1 | 21388757 | LOC_Os01g38180 |
