Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1334-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1334-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 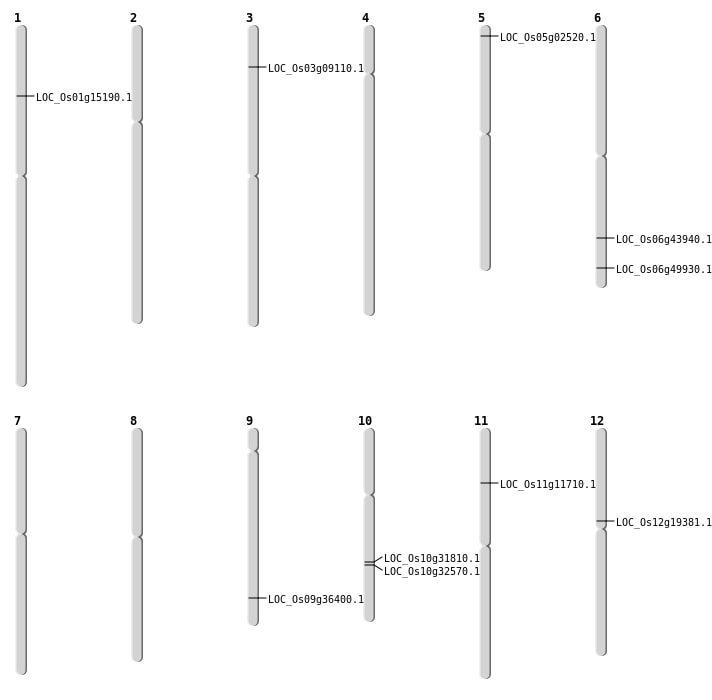
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 39771447 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 8516017 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g15190 |
| SBS | Chr10 | 12740125 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 15131612 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 2331198 | A-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 3230037 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 14918929 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 27754604 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 23123639 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 28043010 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 31271858 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 9649263 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 29922948 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 831402 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 831403 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 874112 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g02520 |
| SBS | Chr5 | 8850798 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 19946846 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 23825629 | G-C | HET | INTRON | |
| SBS | Chr6 | 29881940 | T-G | HET | INTRON | |
| SBS | Chr6 | 30294196 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 2375067 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 26088604 | C-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 26545715 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 19662077 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 21712900 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 7135122 | C-T | HOMO | SYNONYMOUS_CODING |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 2256767 | 2256786 | 19 | |
| Deletion | Chr3 | 4740606 | 4740607 | 1 | LOC_Os03g09110 |
| Deletion | Chr7 | 11063740 | 11063741 | 1 | |
| Deletion | Chr12 | 11275178 | 11275179 | 1 | |
| Deletion | Chr3 | 13639376 | 13639378 | 2 | |
| Deletion | Chr3 | 14843358 | 14843362 | 4 | |
| Deletion | Chr9 | 15474133 | 15474134 | 1 | |
| Deletion | Chr10 | 16677328 | 16677336 | 8 | LOC_Os10g31810 |
| Deletion | Chr10 | 17066412 | 17067787 | 1375 | LOC_Os10g32570 |
| Deletion | Chr10 | 18100909 | 18100915 | 6 | |
| Deletion | Chr9 | 19586329 | 19586334 | 5 | |
| Deletion | Chr6 | 26473366 | 26473377 | 11 | LOC_Os06g43940 |
| Deletion | Chr2 | 29603613 | 29603655 | 42 |
Insertions: 8
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 6435708 | 6459680 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 2744477 | Chr1 | 19170028 | |
| Translocation | Chr12 | 11272986 | Chr11 | 9861376 | LOC_Os12g19381 |
| Translocation | Chr3 | 16632910 | Chr2 | 33429412 | |
| Translocation | Chr10 | 17066421 | Chr7 | 8650187 | LOC_Os10g32570 |
| Translocation | Chr10 | 17067771 | Chr7 | 8650186 | |
| Translocation | Chr12 | 20116853 | Chr11 | 6439248 | |
| Translocation | Chr12 | 20116880 | Chr11 | 6510125 | LOC_Os11g11710 |
| Translocation | Chr9 | 21005583 | Chr6 | 30225341 | 2 |
