Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1338-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1338-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 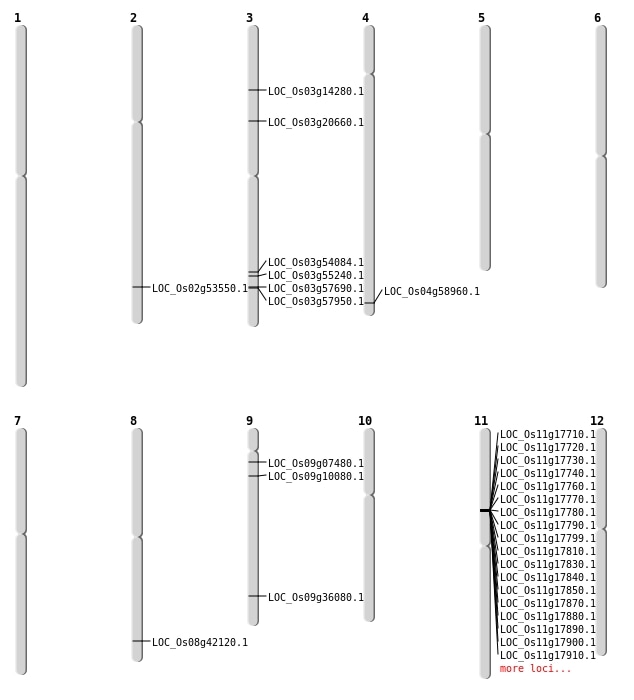
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14108368 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 14108369 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 19389009 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 11477660 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 17736481 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 21437792 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 22547792 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 23275365 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 23768054 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g39870 |
| SBS | Chr11 | 3669565 | C-T | HET | INTRON | |
| SBS | Chr12 | 10271759 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 16261393 | C-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 22100679 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 874106 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 32764606 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g53550 |
| SBS | Chr2 | 35900340 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 23072232 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 25680450 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 26511889 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 30933479 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 31008655 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g54084 |
| SBS | Chr3 | 8166505 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr5 | 17903200 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 24306956 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 10620240 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 12121321 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 5856267 | A-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 13898932 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 14756658 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 20775911 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g36080 |
| SBS | Chr9 | 324318 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 3949170 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 5495450 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g10080 |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 1938975 | 1938992 | 17 | |
| Deletion | Chr9 | 3747220 | 3748268 | 1048 | LOC_Os09g07480 |
| Deletion | Chr11 | 4167462 | 4167463 | 1 | |
| Deletion | Chr5 | 5483891 | 5483892 | 1 | |
| Deletion | Chr3 | 7778094 | 7778110 | 16 | LOC_Os03g14280 |
| Deletion | Chr11 | 9886001 | 9976000 | 90000 | 11 |
| Deletion | Chr11 | 9986001 | 10228000 | 242000 | 26 |
| Deletion | Chr11 | 10243853 | 11216783 | 972930 | 121 |
| Deletion | Chr6 | 10403159 | 10403174 | 15 | |
| Deletion | Chr3 | 11686964 | 11686968 | 4 | LOC_Os03g20660 |
| Deletion | Chr4 | 12679891 | 12679892 | 1 | |
| Deletion | Chr9 | 12944519 | 12945040 | 521 | |
| Deletion | Chr9 | 13482639 | 13482647 | 8 | |
| Deletion | Chr3 | 14763969 | 14763970 | 1 | |
| Deletion | Chr9 | 18793169 | 18793170 | 1 | |
| Deletion | Chr1 | 19434008 | 19434009 | 1 | |
| Deletion | Chr12 | 22243499 | 22243501 | 2 | |
| Deletion | Chr5 | 22707407 | 22707420 | 13 | |
| Deletion | Chr6 | 23034634 | 23034635 | 1 | |
| Deletion | Chr5 | 23469251 | 23470589 | 1338 | |
| Deletion | Chr7 | 23897713 | 23897718 | 5 | |
| Deletion | Chr1 | 24490224 | 24490225 | 1 | |
| Deletion | Chr3 | 30117451 | 30117453 | 2 | |
| Deletion | Chr3 | 31225286 | 31225293 | 7 | |
| Deletion | Chr3 | 32076459 | 32076490 | 31 | |
| Deletion | Chr3 | 32882695 | 32882697 | 2 | LOC_Os03g57690 |
| Deletion | Chr3 | 33014030 | 33014038 | 8 | LOC_Os03g57950 |
| Deletion | Chr4 | 35074674 | 35074677 | 3 | LOC_Os04g58960 |
| Deletion | Chr1 | 36787753 | 36787760 | 7 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 30507248 | 30507249 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 31108298 | 31441908 | LOC_Os03g55240 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 7698801 | Chr8 | 26615553 | LOC_Os08g42120 |
| Translocation | Chr6 | 26619420 | Chr5 | 23470583 | |
| Translocation | Chr6 | 26619428 | Chr5 | 23469261 |
