Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1349-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1349-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 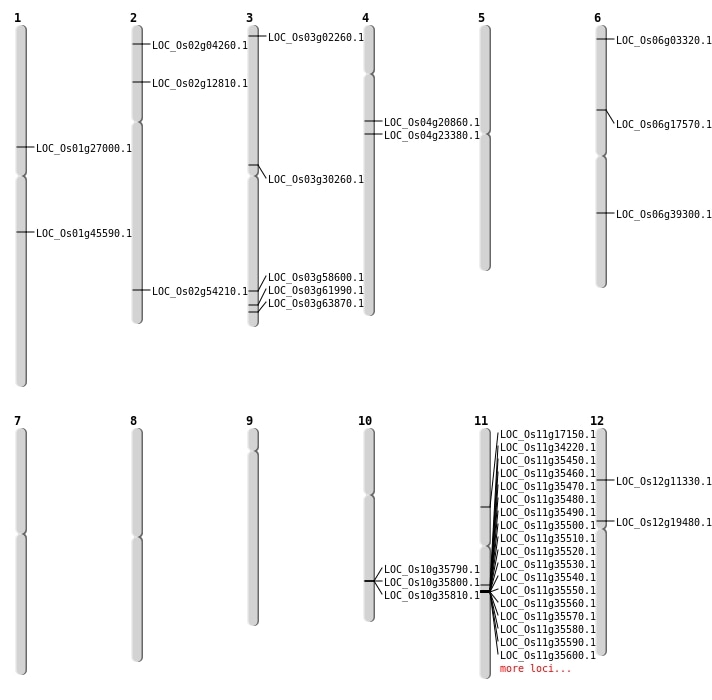
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16138711 | T-A | HET | INTRON | |
| SBS | Chr1 | 27495810 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 18124939 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3197149 | G-A | HET | INTRON | |
| SBS | Chr10 | 6964070 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 25579197 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 11332701 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g19480 |
| SBS | Chr2 | 14327473 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 22912790 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 28372468 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 32956318 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 6727257 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g12810 |
| SBS | Chr3 | 31327199 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 31327206 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 33375057 | A-T | HET | STOP_GAINED | LOC_Os03g58600 |
| SBS | Chr3 | 5867704 | A-G | HET | INTRON | |
| SBS | Chr3 | 767189 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g02260 |
| SBS | Chr4 | 11711927 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g20860 |
| SBS | Chr4 | 13369579 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g23380 |
| SBS | Chr4 | 14544461 | C-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 22190800 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 27200736 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 3999337 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 4088225 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 19910565 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 9185774 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 8822829 | T-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 1795707 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 11168901 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 17516420 | C-A | HOMO | INTRON | |
| SBS | Chr9 | 4233020 | C-T | HOMO | INTERGENIC |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 427528 | 427529 | 1 | |
| Deletion | Chr2 | 1881952 | 1881953 | 1 | LOC_Os02g04260 |
| Deletion | Chr5 | 2126492 | 2126502 | 10 | |
| Deletion | Chr12 | 4803044 | 4803051 | 7 | |
| Deletion | Chr5 | 5618088 | 5618091 | 3 | |
| Deletion | Chr12 | 6109339 | 6109352 | 13 | LOC_Os12g11330 |
| Deletion | Chr12 | 6636565 | 6636566 | 1 | |
| Deletion | Chr11 | 9524195 | 9524210 | 15 | LOC_Os11g17150 |
| Deletion | Chr8 | 10177457 | 10177458 | 1 | |
| Deletion | Chr6 | 10198772 | 10198773 | 1 | LOC_Os06g17570 |
| Deletion | Chr9 | 12643950 | 12643953 | 3 | |
| Deletion | Chr12 | 14289911 | 14289913 | 2 | |
| Deletion | Chr1 | 15044169 | 15044170 | 1 | LOC_Os01g27000 |
| Deletion | Chr8 | 15970751 | 15970752 | 1 | |
| Deletion | Chr7 | 16971002 | 16971039 | 37 | |
| Deletion | Chr3 | 17263156 | 17263158 | 2 | LOC_Os03g30260 |
| Deletion | Chr12 | 17565404 | 17565414 | 10 | |
| Deletion | Chr10 | 19141357 | 19147682 | 6325 | 3 |
| Deletion | Chr6 | 19910571 | 19910574 | 3 | |
| Deletion | Chr3 | 20047812 | 20047831 | 19 | |
| Deletion | Chr1 | 20155444 | 20155445 | 1 | |
| Deletion | Chr11 | 20772001 | 21177000 | 405000 | 58 |
| Deletion | Chr8 | 20877890 | 20877903 | 13 | |
| Deletion | Chr12 | 21094169 | 21094182 | 13 | |
| Deletion | Chr3 | 22092643 | 22092644 | 1 | |
| Deletion | Chr8 | 23297357 | 23297358 | 1 | |
| Deletion | Chr6 | 23319307 | 23323795 | 4488 | LOC_Os06g39300 |
| Deletion | Chr1 | 25895283 | 25895286 | 3 | LOC_Os01g45590 |
| Deletion | Chr3 | 35032123 | 35032125 | 2 | |
| Deletion | Chr3 | 35134800 | 35134802 | 2 | LOC_Os03g61990 |
| Deletion | Chr3 | 36095415 | 36095430 | 15 | LOC_Os03g63870 |
| Deletion | Chr1 | 36596220 | 36596248 | 28 | |
| Deletion | Chr1 | 39265569 | 39265571 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 24620953 | 24620970 | 18 | |
| Insertion | Chr2 | 35559918 | 35559920 | 3 | |
| Insertion | Chr4 | 18177773 | 18177773 | 1 |
No Inversion
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 1266437 | Chr2 | 33237978 | 2 |
| Translocation | Chr11 | 12913942 | Chr6 | 23319388 | |
| Translocation | Chr11 | 12913947 | Chr6 | 23323813 | LOC_Os06g39300 |
| Translocation | Chr10 | 19141361 | Chr5 | 6062585 | |
| Translocation | Chr10 | 19147676 | Chr5 | 6062548 | LOC_Os10g35800 |
| Translocation | Chr11 | 20037514 | Chr6 | 1427715 | LOC_Os11g34220 |
| Translocation | Chr11 | 20039628 | Chr6 | 1427708 |
