Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1504-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1504-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 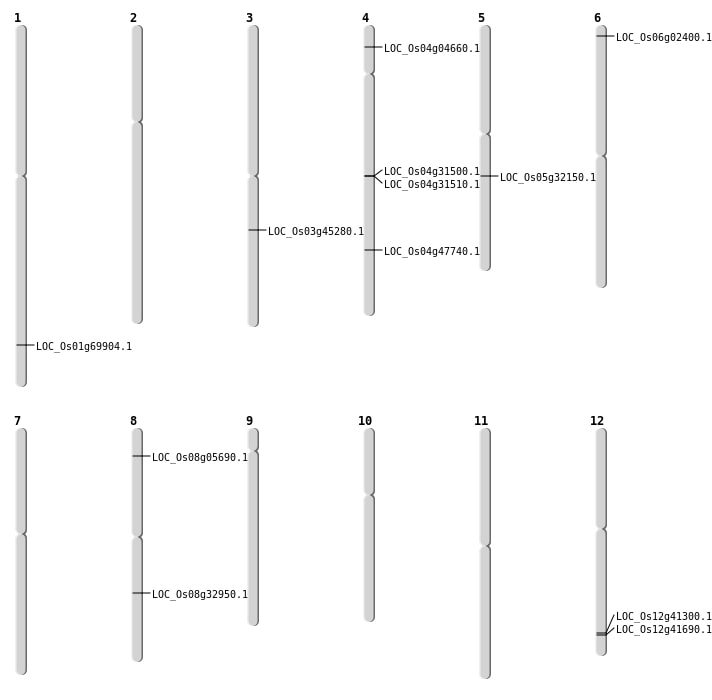
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 13447174 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 19993973 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 5737618 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 11121747 | T-A | HET | INTRON | |
| SBS | Chr11 | 1144088 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 16476863 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 17692818 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 13501698 | G-A | HET | INTRON | |
| SBS | Chr12 | 21665897 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 35310862 | C-T | HET | INTRON | |
| SBS | Chr2 | 4413699 | A-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr3 | 25563850 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g45280 |
| SBS | Chr3 | 28858259 | C-A | HET | INTRON | |
| SBS | Chr3 | 4450809 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 4450811 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 13121429 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 18152106 | G-C | HET | INTRON | |
| SBS | Chr5 | 13518996 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 16597273 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 22643134 | A-C | HET | INTRON | |
| SBS | Chr5 | 22788001 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 501319 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 15276801 | A-G | HET | INTRON | |
| SBS | Chr6 | 28790648 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr7 | 10151327 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 17087974 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 10947400 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 12771736 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 18401683 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 1087505 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 9421770 | C-T | Homo | INTRON |
Deletions: 43
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 782246 | 782252 | 6 | |
| Deletion | Chr6 | 828576 | 828579 | 3 | LOC_Os06g02400 |
| Deletion | Chr12 | 1356536 | 1356549 | 13 | |
| Deletion | Chr4 | 2243416 | 2243440 | 24 | LOC_Os04g04660 |
| Deletion | Chr5 | 2339784 | 2339785 | 1 | |
| Deletion | Chr6 | 3425266 | 3425283 | 17 | |
| Deletion | Chr4 | 3440427 | 3440442 | 15 | |
| Deletion | Chr11 | 4037737 | 4037741 | 4 | |
| Deletion | Chr9 | 4712866 | 4712867 | 1 | |
| Deletion | Chr7 | 5014058 | 5014059 | 1 | |
| Deletion | Chr11 | 5813421 | 5813422 | 1 | |
| Deletion | Chr8 | 6471242 | 6471243 | 1 | |
| Deletion | Chr10 | 8537419 | 8537432 | 13 | |
| Deletion | Chr3 | 11165795 | 11165796 | 1 | |
| Deletion | Chr5 | 11580439 | 11580440 | 1 | |
| Deletion | Chr9 | 11631521 | 11631522 | 1 | |
| Deletion | Chr11 | 12121862 | 12121865 | 3 | |
| Deletion | Chr12 | 12918934 | 12918935 | 1 | |
| Deletion | Chr8 | 13492441 | 13492451 | 10 | |
| Deletion | Chr1 | 13729515 | 13729620 | 105 | |
| Deletion | Chr1 | 14221755 | 14221765 | 10 | |
| Deletion | Chr1 | 17343714 | 17343715 | 1 | |
| Deletion | Chr6 | 18334383 | 18334384 | 1 | |
| Deletion | Chr4 | 18831260 | 18842850 | 11590 | 2 |
| Deletion | Chr6 | 19601085 | 19601087 | 2 | |
| Deletion | Chr10 | 19994009 | 19994016 | 7 | |
| Deletion | Chr5 | 21023064 | 21023068 | 4 | |
| Deletion | Chr12 | 22900096 | 22900097 | 1 | |
| Deletion | Chr7 | 23067593 | 23068626 | 1033 | |
| Deletion | Chr12 | 23447939 | 23447959 | 20 | |
| Deletion | Chr7 | 24032521 | 24032534 | 13 | |
| Deletion | Chr12 | 24312050 | 24312052 | 2 | |
| Deletion | Chr8 | 25088502 | 25088512 | 10 | |
| Deletion | Chr12 | 25613426 | 25613433 | 7 | LOC_Os12g41300 |
| Deletion | Chr12 | 25807763 | 25807765 | 2 | LOC_Os12g41690 |
| Deletion | Chr6 | 26711992 | 26711993 | 1 | |
| Deletion | Chr4 | 28329054 | 28329068 | 14 | LOC_Os04g47740 |
| Deletion | Chr5 | 28606984 | 28606988 | 4 | |
| Deletion | Chr6 | 29724518 | 29724519 | 1 | |
| Deletion | Chr2 | 30280023 | 30280024 | 1 | |
| Deletion | Chr4 | 33225772 | 33225773 | 1 | |
| Deletion | Chr1 | 40391688 | 40391694 | 6 | LOC_Os01g69904 |
| Deletion | Chr1 | 40820405 | 40820421 | 16 |
Insertions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 13350424 | 13350444 | 21 | |
| Insertion | Chr1 | 21317022 | 21317022 | 1 | |
| Insertion | Chr1 | 39666745 | 39666792 | 48 | |
| Insertion | Chr11 | 22360199 | 22360221 | 23 | |
| Insertion | Chr11 | 8175447 | 8175513 | 67 | |
| Insertion | Chr3 | 14279840 | 14279841 | 2 | |
| Insertion | Chr3 | 6180775 | 6180775 | 1 | |
| Insertion | Chr4 | 27963828 | 27963828 | 1 | |
| Insertion | Chr5 | 18743356 | 18743373 | 18 | LOC_Os05g32150 |
| Insertion | Chr5 | 20708415 | 20708420 | 6 | |
| Insertion | Chr6 | 24690339 | 24690344 | 6 | |
| Insertion | Chr7 | 11792515 | 11792516 | 2 | |
| Insertion | Chr7 | 9166822 | 9166823 | 2 | |
| Insertion | Chr8 | 15507494 | 15507494 | 1 | |
| Insertion | Chr8 | 20336766 | 20336779 | 14 | |
| Insertion | Chr8 | 20447908 | 20447978 | 71 | LOC_Os08g32950 |
| Insertion | Chr8 | 25678116 | 25678118 | 3 | |
| Insertion | Chr8 | 3053677 | 3053677 | 1 | LOC_Os08g05690 |
| Insertion | Chr8 | 3168958 | 3168959 | 2 |
No Inversion
No Translocation
