Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1517-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1517-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 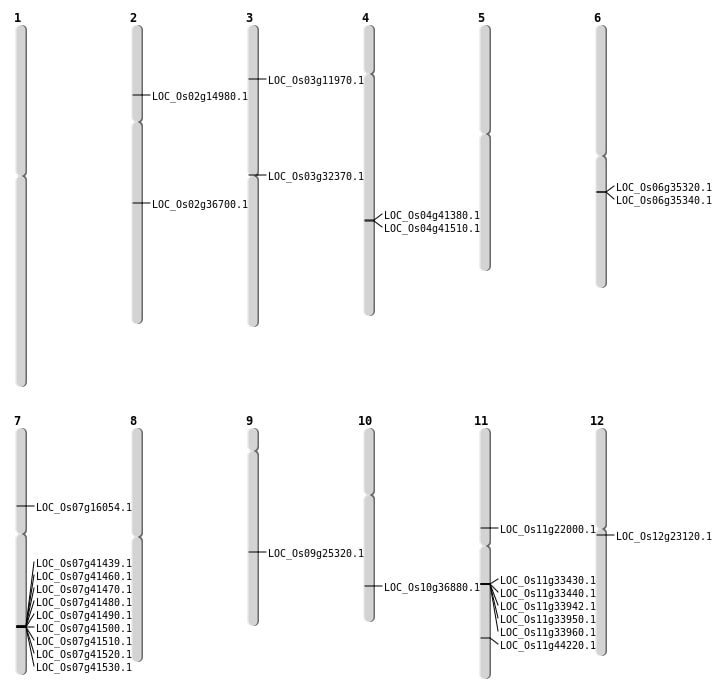
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22410212 | A-G | Homo | INTRON | |
| SBS | Chr10 | 12674791 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 1424915 | C-G | Homo | INTERGENIC | |
| SBS | Chr10 | 19758875 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g36880 |
| SBS | Chr10 | 22905633 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 23072495 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 12645430 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g22000 |
| SBS | Chr11 | 23638050 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 24708058 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 7987108 | T-C | Homo | SYNONYMOUS_CODING | |
| SBS | Chr12 | 24855298 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 12755498 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 19175765 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 30403031 | G-T | HET | INTRON | |
| SBS | Chr2 | 7930367 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 20463986 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 6922742 | T-A | Homo | INTERGENIC | |
| SBS | Chr4 | 27269884 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 12227293 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 17393812 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 20757363 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 29830042 | C-T | HET | INTRON | |
| SBS | Chr7 | 1129371 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 12856830 | T-A | Homo | INTERGENIC | |
| SBS | Chr7 | 25276296 | C-T | HET | INTRON | |
| SBS | Chr7 | 26395818 | A-G | Homo | INTRON | |
| SBS | Chr7 | 26395819 | G-A | Homo | INTRON | |
| SBS | Chr8 | 1618445 | A-T | Homo | INTERGENIC | |
| SBS | Chr8 | 273151 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr9 | 12462740 | G-A | HET | INTERGENIC |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 3478370 | 3478371 | 1 | |
| Deletion | Chr12 | 4664594 | 4664611 | 17 | |
| Deletion | Chr3 | 6276898 | 6276901 | 3 | LOC_Os03g11970 |
| Deletion | Chr10 | 6407832 | 6407834 | 2 | |
| Deletion | Chr10 | 7139427 | 7139428 | 1 | |
| Deletion | Chr6 | 7544076 | 7544154 | 78 | |
| Deletion | Chr11 | 7849046 | 7849048 | 2 | |
| Deletion | Chr2 | 7933165 | 7933170 | 5 | |
| Deletion | Chr2 | 8361458 | 8362049 | 591 | LOC_Os02g14980 |
| Deletion | Chr7 | 9359689 | 9359690 | 1 | LOC_Os07g16054 |
| Deletion | Chr9 | 12252232 | 12252233 | 1 | |
| Deletion | Chr6 | 12448801 | 12448802 | 1 | |
| Deletion | Chr12 | 13071960 | 13071961 | 1 | LOC_Os12g23120 |
| Deletion | Chr5 | 13786242 | 13786243 | 1 | |
| Deletion | Chr3 | 13968958 | 13968960 | 2 | |
| Deletion | Chr6 | 14715585 | 14715586 | 1 | |
| Deletion | Chr12 | 15730470 | 15730471 | 1 | |
| Deletion | Chr12 | 17255243 | 17255246 | 3 | |
| Deletion | Chr3 | 18516914 | 18516917 | 3 | LOC_Os03g32370 |
| Deletion | Chr11 | 19807001 | 19839000 | 32000 | 5 |
| Deletion | Chr6 | 20589674 | 20608157 | 18483 | 2 |
| Deletion | Chr2 | 22146849 | 22146851 | 2 | LOC_Os02g36700 |
| Deletion | Chr11 | 22255634 | 22255635 | 1 | |
| Deletion | Chr2 | 22271287 | 22271303 | 16 | |
| Deletion | Chr9 | 22502344 | 22502349 | 5 | |
| Deletion | Chr5 | 23953524 | 23953538 | 14 | |
| Deletion | Chr7 | 24831001 | 24897000 | 66000 | 9 |
| Deletion | Chr6 | 26415083 | 26415089 | 6 | |
| Deletion | Chr11 | 26716027 | 26716044 | 17 | LOC_Os11g44220 |
| Deletion | Chr3 | 29280008 | 29280013 | 5 | |
| Deletion | Chr3 | 31900589 | 31900590 | 1 | |
| Deletion | Chr3 | 32506504 | 32506512 | 8 |
Insertions: 18
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 24543341 | 24619753 | 2 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 15161379 | Chr2 | 4449264 | LOC_Os09g25320 |
| Translocation | Chr4 | 24619796 | Chr2 | 14658127 | LOC_Os04g41510 |
