Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1577-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1577-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 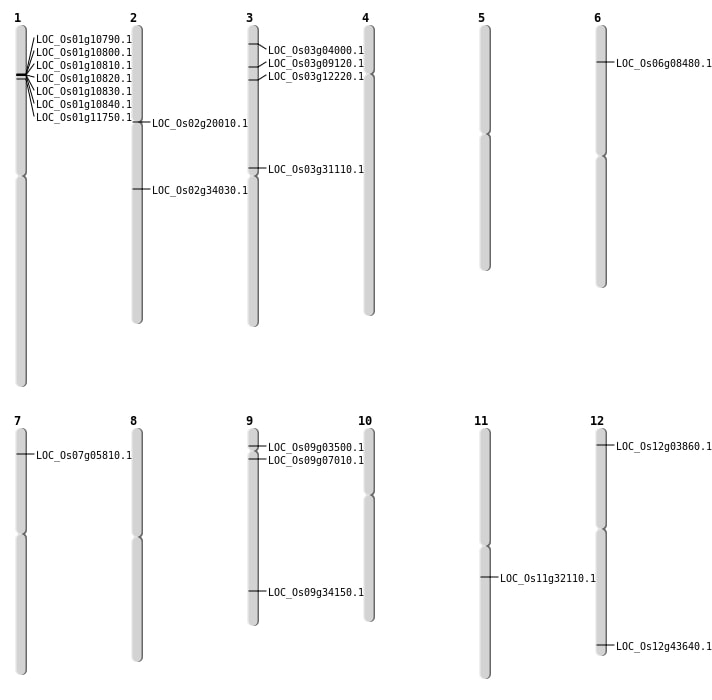
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19429172 | C-T | HET | INTRON | |
| SBS | Chr1 | 40126191 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 16672545 | C-A | HET | INTRON | |
| SBS | Chr10 | 9301280 | G-A | HET | INTRON | |
| SBS | Chr11 | 18034811 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 18926774 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 24322522 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 12317141 | G-T | HET | INTRON | |
| SBS | Chr3 | 11447194 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 1829114 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 23123639 | G-A | Homo | INTERGENIC | |
| SBS | Chr3 | 27362819 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 29447520 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 29533626 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 34929955 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 4040670 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 4160217 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 4362567 | G-A | HET | INTRON | |
| SBS | Chr3 | 5792500 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 5995069 | T-G | HET | INTRON | |
| SBS | Chr3 | 6411356 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g12220 |
| SBS | Chr3 | 7522578 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 9593595 | C-T | HET | INTRON | |
| SBS | Chr4 | 16334172 | C-T | HET | INTRON | |
| SBS | Chr4 | 1836475 | T-A | HET | INTRON | |
| SBS | Chr4 | 31529212 | G-A | HET | INTRON | |
| SBS | Chr6 | 15519406 | C-G | Homo | INTERGENIC | |
| SBS | Chr6 | 22063462 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 24743920 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 9308339 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 4172822 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 4172823 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 4172824 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 12324413 | C-T | Homo | INTERGENIC | |
| SBS | Chr9 | 10455630 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 10998745 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 1728165 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g03500 |
| SBS | Chr9 | 18101066 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 18101068 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 3224498 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 8482786 | C-T | HET | SYNONYMOUS_CODING |
Deletions: 39
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 708509 | 708515 | 6 | |
| Deletion | Chr3 | 1825091 | 1825104 | 13 | LOC_Os03g04000 |
| Deletion | Chr7 | 2795860 | 2795872 | 12 | LOC_Os07g05810 |
| Deletion | Chr9 | 3400981 | 3401516 | 535 | LOC_Os09g07010 |
| Deletion | Chr3 | 3694948 | 3694950 | 2 | |
| Deletion | Chr6 | 4171864 | 4171878 | 14 | LOC_Os06g08480 |
| Deletion | Chr3 | 4479338 | 4479347 | 9 | |
| Deletion | Chr3 | 4749517 | 4749518 | 1 | LOC_Os03g09120 |
| Deletion | Chr1 | 5286461 | 5286462 | 1 | |
| Deletion | Chr1 | 5763419 | 5782119 | 18700 | 6 |
| Deletion | Chr3 | 8910585 | 8910593 | 8 | |
| Deletion | Chr3 | 9714244 | 9714246 | 2 | |
| Deletion | Chr6 | 9762737 | 9762738 | 1 | |
| Deletion | Chr3 | 10847158 | 10847184 | 26 | |
| Deletion | Chr3 | 10883674 | 10883676 | 2 | |
| Deletion | Chr2 | 11781594 | 11781596 | 2 | LOC_Os02g20010 |
| Deletion | Chr9 | 12239911 | 12239913 | 2 | |
| Deletion | Chr10 | 13331877 | 13331880 | 3 | |
| Deletion | Chr5 | 13712723 | 13712724 | 1 | |
| Deletion | Chr9 | 14761264 | 14761265 | 1 | |
| Deletion | Chr4 | 15219036 | 15219037 | 1 | |
| Deletion | Chr2 | 15621705 | 15621706 | 1 | |
| Deletion | Chr6 | 16334873 | 16334883 | 10 | |
| Deletion | Chr11 | 18937849 | 18937865 | 16 | LOC_Os11g32110 |
| Deletion | Chr9 | 20159233 | 20159234 | 1 | LOC_Os09g34150 |
| Deletion | Chr2 | 20324307 | 20324322 | 15 | LOC_Os02g34030 |
| Deletion | Chr9 | 20563755 | 20563756 | 1 | |
| Deletion | Chr12 | 22889707 | 22889711 | 4 | |
| Deletion | Chr2 | 23103954 | 23103956 | 2 | |
| Deletion | Chr7 | 23110573 | 23110582 | 9 | |
| Deletion | Chr8 | 23528681 | 23528708 | 27 | |
| Deletion | Chr5 | 25071762 | 25071768 | 6 | |
| Deletion | Chr6 | 26386188 | 26386190 | 2 | |
| Deletion | Chr6 | 26684687 | 26684709 | 22 | |
| Deletion | Chr1 | 27071934 | 27071952 | 18 | |
| Deletion | Chr1 | 28532120 | 28532140 | 20 | |
| Deletion | Chr7 | 28970120 | 28970121 | 1 | |
| Deletion | Chr1 | 38982744 | 38982761 | 17 | |
| Deletion | Chr1 | 41684598 | 41684599 | 1 |
Insertions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 1586845 | 1586847 | 3 | LOC_Os12g03860 |
| Insertion | Chr12 | 27110447 | 27110518 | 72 | LOC_Os12g43640 |
| Insertion | Chr2 | 12658737 | 12658737 | 1 | |
| Insertion | Chr3 | 30630610 | 30630611 | 2 | |
| Insertion | Chr3 | 536553 | 536562 | 10 | |
| Insertion | Chr4 | 33496912 | 33496912 | 1 | |
| Insertion | Chr4 | 524552 | 524555 | 4 | |
| Insertion | Chr5 | 20925680 | 20925680 | 1 | |
| Insertion | Chr5 | 28539576 | 28539585 | 10 | |
| Insertion | Chr5 | 7022961 | 7022972 | 12 | |
| Insertion | Chr5 | 9868725 | 9868725 | 1 | |
| Insertion | Chr6 | 26142892 | 26142899 | 8 | |
| Insertion | Chr7 | 10121392 | 10121435 | 44 | |
| Insertion | Chr7 | 1501250 | 1501253 | 4 | |
| Insertion | Chr8 | 14433173 | 14433173 | 1 | |
| Insertion | Chr8 | 22508646 | 22508646 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 5780919 | 6362487 | LOC_Os01g11750 |
| Inversion | Chr3 | 17720780 | 19449844 | LOC_Os03g31110 |
No Translocation
