Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1596-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1596-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 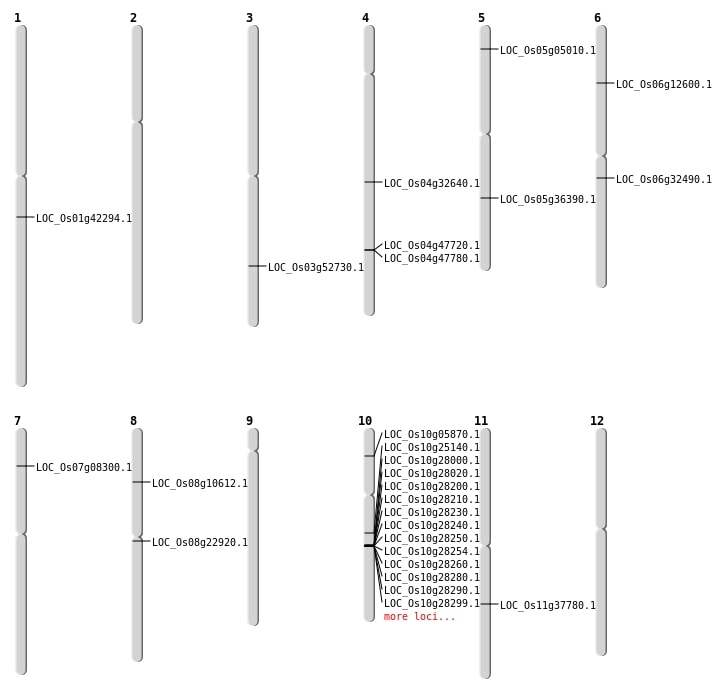
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 141441 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr1 | 2836818 | C-T | HET | INTRON | |
| SBS | Chr1 | 38672184 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 42401251 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 43015889 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 12981280 | C-T | HET | INTRON | |
| SBS | Chr11 | 13029333 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 17670083 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 22369472 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g37780 |
| SBS | Chr11 | 23321936 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 9085721 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 21024179 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 16356306 | C-A | HET | INTRON | |
| SBS | Chr3 | 24323657 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 29218680 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 31941483 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 32574595 | G-A | HET | INTRON | |
| SBS | Chr3 | 32574604 | T-A | HET | INTRON | |
| SBS | Chr4 | 13917986 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 5853983 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 17052612 | C-T | HET | INTRON | |
| SBS | Chr5 | 25322312 | C-T | HET | INTRON | |
| SBS | Chr5 | 2716820 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr5 | 2716822 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr5 | 4889555 | A-G | HET | INTRON | |
| SBS | Chr5 | 5963950 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 7915116 | C-T | HET | INTRON | |
| SBS | Chr7 | 11259086 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 7888086 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 11526090 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 1900784 | T-A | HET | UTR_3_PRIME | |
| SBS | Chr9 | 22865355 | C-A | HET | INTRON |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 328997 | 328999 | 2 | |
| Deletion | Chr5 | 2431698 | 2431709 | 11 | LOC_Os05g05010 |
| Deletion | Chr5 | 2540993 | 2541003 | 10 | |
| Deletion | Chr5 | 3048054 | 3048068 | 14 | |
| Deletion | Chr7 | 4252927 | 4252933 | 6 | LOC_Os07g08300 |
| Deletion | Chr9 | 4941817 | 4941818 | 1 | |
| Deletion | Chr10 | 5704857 | 5704859 | 2 | |
| Deletion | Chr8 | 6251498 | 6251508 | 10 | LOC_Os08g10612 |
| Deletion | Chr2 | 6269283 | 6269289 | 6 | |
| Deletion | Chr6 | 6837790 | 6852038 | 14248 | LOC_Os06g12600 |
| Deletion | Chr8 | 9043557 | 9043631 | 74 | |
| Deletion | Chr8 | 9880905 | 9880907 | 2 | |
| Deletion | Chr7 | 11037169 | 11037172 | 3 | |
| Deletion | Chr8 | 13780187 | 13780192 | 5 | LOC_Os08g22920 |
| Deletion | Chr12 | 14521950 | 14521951 | 1 | |
| Deletion | Chr10 | 14641001 | 14749000 | 108000 | 14 |
| Deletion | Chr11 | 15761101 | 15761110 | 9 | |
| Deletion | Chr8 | 16387203 | 16387205 | 2 | |
| Deletion | Chr5 | 17935498 | 17935500 | 2 | |
| Deletion | Chr11 | 18118474 | 18118479 | 5 | |
| Deletion | Chr3 | 18134877 | 18134893 | 16 | |
| Deletion | Chr2 | 18443455 | 18443462 | 7 | |
| Deletion | Chr6 | 18887166 | 18899357 | 12191 | LOC_Os06g32490 |
| Deletion | Chr4 | 19669842 | 19669851 | 9 | LOC_Os04g32640 |
| Deletion | Chr11 | 22376938 | 22376939 | 1 | |
| Deletion | Chr7 | 23689006 | 23689011 | 5 | |
| Deletion | Chr7 | 24537535 | 24537536 | 1 | |
| Deletion | Chr6 | 24774690 | 24774691 | 1 | |
| Deletion | Chr2 | 26412755 | 26412757 | 2 | |
| Deletion | Chr8 | 27624862 | 27624864 | 2 | |
| Deletion | Chr4 | 29651040 | 29651045 | 5 | |
| Deletion | Chr3 | 30230241 | 30230249 | 8 | LOC_Os03g52730 |
| Deletion | Chr2 | 30607676 | 30607677 | 1 | |
| Deletion | Chr2 | 32808356 | 32808358 | 2 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23976162 | 23976163 | 2 | LOC_Os01g42294 |
| Insertion | Chr10 | 2820946 | 2820946 | 1 | |
| Insertion | Chr3 | 1185321 | 1185322 | 2 | |
| Insertion | Chr3 | 36062573 | 36062586 | 14 | |
| Insertion | Chr4 | 27444221 | 27444223 | 3 | |
| Insertion | Chr5 | 46709 | 46717 | 9 | |
| Insertion | Chr7 | 8574640 | 8574640 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 2582452 | 2964188 | LOC_Os10g05870 |
| Inversion | Chr10 | 2964185 | 3165880 | LOC_Os10g05870 |
| Inversion | Chr10 | 3012394 | 12864006 | |
| Inversion | Chr10 | 3012438 | 3165953 | |
| Inversion | Chr10 | 14507328 | 14748895 | 2 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 3010943 | Chr4 | 24091624 | |
| Translocation | Chr10 | 3011155 | Chr4 | 24091616 | |
| Translocation | Chr10 | 3011160 | Chr4 | 20624036 | |
| Translocation | Chr10 | 3011987 | Chr4 | 20624009 | |
| Translocation | Chr10 | 3012277 | Chr4 | 28316569 | LOC_Os04g47720 |
| Translocation | Chr10 | 12864049 | Chr4 | 28316570 | LOC_Os04g47720 |
| Translocation | Chr10 | 12980885 | Chr4 | 28342343 | 2 |
| Translocation | Chr10 | 14540551 | Chr5 | 21535561 | 2 |
