Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1624-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1624-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 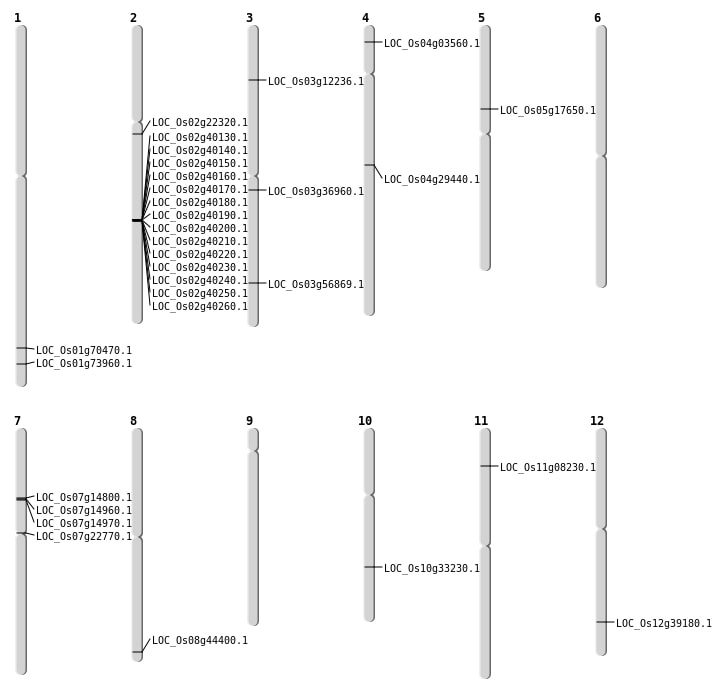
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 42929885 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 6603405 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 13926058 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 17432821 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g33230 |
| SBS | Chr10 | 5914971 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 22880010 | T-A | HET | INTRON | |
| SBS | Chr11 | 28533380 | G-C | Homo | INTERGENIC | |
| SBS | Chr11 | 4320944 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g08230 |
| SBS | Chr12 | 13792524 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 24043240 | G-A | Homo | INTERGENIC | |
| SBS | Chr2 | 8709671 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 19852552 | T-G | HET | INTRON | |
| SBS | Chr3 | 32396392 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g56869 |
| SBS | Chr3 | 7061228 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 8738022 | C-T | HET | INTRON | |
| SBS | Chr4 | 1553940 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g03560 |
| SBS | Chr4 | 17006703 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 10158833 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17650 |
| SBS | Chr5 | 24824624 | T-G | Homo | INTERGENIC | |
| SBS | Chr5 | 6709328 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 7345320 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 11481745 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 12403177 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 16604359 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 17714712 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 21056730 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 21406092 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 26844200 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 5808453 | A-G | HET | INTRON | |
| SBS | Chr7 | 8563563 | A-G | Homo | SYNONYMOUS_CODING | |
| SBS | Chr8 | 10757764 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 16616804 | C-T | HET | INTRON | |
| SBS | Chr8 | 16616805 | C-T | HET | INTRON | |
| SBS | Chr8 | 27473254 | T-C | Homo | INTERGENIC | |
| SBS | Chr8 | 9660104 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 7951684 | G-C | HET | INTERGENIC |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 626005 | 626006 | 1 | |
| Deletion | Chr12 | 1251785 | 1251786 | 1 | |
| Deletion | Chr3 | 2789662 | 2789686 | 24 | |
| Deletion | Chr9 | 2960840 | 2960842 | 2 | |
| Deletion | Chr2 | 4167178 | 4167185 | 7 | LOC_Os02g07960 |
| Deletion | Chr7 | 4450071 | 4450109 | 38 | |
| Deletion | Chr12 | 4613578 | 4613579 | 1 | |
| Deletion | Chr1 | 6236054 | 6236065 | 11 | |
| Deletion | Chr3 | 6426888 | 6426908 | 20 | LOC_Os03g12236 |
| Deletion | Chr7 | 8452729 | 8452730 | 1 | LOC_Os07g14800 |
| Deletion | Chr10 | 9020005 | 9020008 | 3 | |
| Deletion | Chr1 | 9105570 | 9105571 | 1 | |
| Deletion | Chr11 | 9328077 | 9328138 | 61 | |
| Deletion | Chr9 | 11356385 | 11356386 | 1 | |
| Deletion | Chr1 | 12698489 | 12698563 | 74 | |
| Deletion | Chr7 | 12848499 | 12848501 | 2 | LOC_Os07g22770 |
| Deletion | Chr2 | 13330923 | 13330925 | 2 | LOC_Os02g22320 |
| Deletion | Chr4 | 14979650 | 14979651 | 1 | |
| Deletion | Chr1 | 15549394 | 15549425 | 31 | |
| Deletion | Chr8 | 15928568 | 15928934 | 366 | |
| Deletion | Chr9 | 19021958 | 19021963 | 5 | |
| Deletion | Chr9 | 19451399 | 19451442 | 43 | |
| Deletion | Chr3 | 20501617 | 20501634 | 17 | LOC_Os03g36960 |
| Deletion | Chr5 | 22353047 | 22353048 | 1 | |
| Deletion | Chr3 | 23834616 | 23834617 | 1 | |
| Deletion | Chr12 | 24120936 | 24120958 | 22 | LOC_Os12g39180 |
| Deletion | Chr7 | 24281799 | 24281808 | 9 | |
| Deletion | Chr2 | 24293001 | 24373000 | 80000 | 14 |
| Deletion | Chr12 | 24805609 | 24805610 | 1 | |
| Deletion | Chr12 | 25225944 | 25225972 | 28 | |
| Deletion | Chr6 | 27331199 | 27331200 | 1 | |
| Deletion | Chr3 | 28911787 | 28911788 | 1 | |
| Deletion | Chr1 | 32876233 | 32876234 | 1 | |
| Deletion | Chr1 | 40813821 | 40813824 | 3 | LOC_Os01g70470 |
| Deletion | Chr1 | 42848079 | 42848084 | 5 | LOC_Os01g73960 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 14631745 | 14631746 | 2 | |
| Insertion | Chr2 | 4781090 | 4781090 | 1 | |
| Insertion | Chr3 | 11487828 | 11487829 | 2 | |
| Insertion | Chr4 | 17505265 | 17505288 | 24 | LOC_Os04g29440 |
| Insertion | Chr6 | 3986112 | 3986177 | 66 | |
| Insertion | Chr7 | 22480488 | 22480488 | 1 | |
| Insertion | Chr8 | 15872777 | 15872778 | 2 | |
| Insertion | Chr8 | 17321864 | 17321864 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 8582785 | 8590917 | 2 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 9940221 | Chr7 | 8590927 | LOC_Os07g14970 |
| Translocation | Chr9 | 13582068 | Chr3 | 10613253 | |
| Translocation | Chr6 | 13699050 | Chr1 | 2503882 | |
| Translocation | Chr8 | 27940341 | Chr2 | 10385056 | LOC_Os08g44400 |
| Translocation | Chr8 | 27940726 | Chr2 | 10385137 | LOC_Os08g44400 |
| Translocation | Chr8 | 28218761 | Chr2 | 13833675 | |
| Translocation | Chr8 | 28219680 | Chr2 | 13833709 |
