Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1655-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1655-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 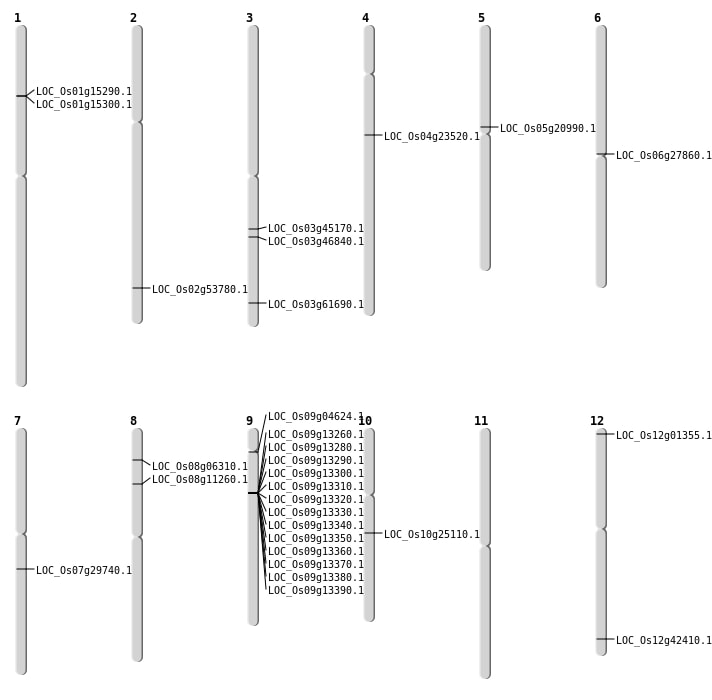
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 12943044 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 14765702 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 10378174 | G-A | Homo | INTERGENIC | |
| SBS | Chr11 | 14754728 | C-T | Homo | INTERGENIC | |
| SBS | Chr11 | 20833182 | A-C | Homo | UTR_3_PRIME | |
| SBS | Chr11 | 2165586 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 8538916 | C-G | Homo | INTRON | |
| SBS | Chr12 | 23269745 | T-C | HET | INTRON | |
| SBS | Chr12 | 3698090 | T-G | HET | INTRON | |
| SBS | Chr2 | 16352402 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 25961271 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 23460090 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 23481040 | T-A | Homo | INTERGENIC | |
| SBS | Chr4 | 15113004 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 32790046 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g55150 |
| SBS | Chr4 | 6667854 | C-G | HET | INTRON | |
| SBS | Chr5 | 13500907 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 3097057 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 16435643 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 20424863 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 20983567 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 17484021 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g29740 |
| SBS | Chr8 | 11002016 | G-A | Homo | INTERGENIC | |
| SBS | Chr8 | 12738816 | T-A | HET | INTRON | |
| SBS | Chr8 | 12738818 | G-A | HET | INTRON | |
| SBS | Chr8 | 15628480 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 17598255 | G-A | Homo | SYNONYMOUS_CODING | |
| SBS | Chr8 | 6620200 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g11260 |
| SBS | Chr8 | 7170581 | C-T | HET | INTRON | |
| SBS | Chr8 | 9601011 | G-A | Homo | INTERGENIC | |
| SBS | Chr9 | 7323076 | G-T | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 212814 | 212845 | 31 | LOC_Os12g01355 |
| Deletion | Chr7 | 1075018 | 1075029 | 11 | |
| Deletion | Chr9 | 2471214 | 2471217 | 3 | LOC_Os09g04624 |
| Deletion | Chr8 | 3482543 | 3482553 | 10 | LOC_Os08g06310 |
| Deletion | Chr5 | 6549539 | 6549540 | 1 | |
| Deletion | Chr9 | 7646001 | 7741000 | 95000 | 13 |
| Deletion | Chr1 | 8554328 | 8561216 | 6888 | 2 |
| Deletion | Chr8 | 8573775 | 8573779 | 4 | |
| Deletion | Chr6 | 11240301 | 11240303 | 2 | |
| Deletion | Chr10 | 11321264 | 11321270 | 6 | |
| Deletion | Chr5 | 11895391 | 11895393 | 2 | |
| Deletion | Chr3 | 12131436 | 12131440 | 4 | |
| Deletion | Chr4 | 13452412 | 13452415 | 3 | LOC_Os04g23520 |
| Deletion | Chr9 | 14422730 | 14422746 | 16 | |
| Deletion | Chr6 | 15782775 | 15782788 | 13 | LOC_Os06g27860 |
| Deletion | Chr1 | 17118821 | 17118824 | 3 | |
| Deletion | Chr10 | 18111841 | 18111852 | 11 | |
| Deletion | Chr7 | 18833320 | 18833321 | 1 | |
| Deletion | Chr1 | 21174926 | 21174938 | 12 | |
| Deletion | Chr4 | 21368078 | 21368079 | 1 | |
| Deletion | Chr2 | 21839456 | 21840286 | 830 | |
| Deletion | Chr2 | 22191555 | 22191563 | 8 | |
| Deletion | Chr3 | 25505733 | 25505734 | 1 | LOC_Os03g45170 |
| Deletion | Chr12 | 26359280 | 26359281 | 1 | LOC_Os12g42410 |
| Deletion | Chr3 | 26499767 | 26499768 | 1 | LOC_Os03g46840 |
| Deletion | Chr7 | 27748554 | 27748555 | 1 | |
| Deletion | Chr6 | 30031947 | 30031959 | 12 | |
| Deletion | Chr4 | 34600183 | 34600186 | 3 | |
| Deletion | Chr1 | 43264101 | 43264108 | 7 |
Insertions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 37561525 | 37561525 | 1 | |
| Insertion | Chr11 | 19615789 | 19615818 | 30 | |
| Insertion | Chr11 | 22634284 | 22634285 | 2 | |
| Insertion | Chr2 | 21191440 | 21191440 | 1 | |
| Insertion | Chr2 | 33915860 | 33915860 | 1 | |
| Insertion | Chr3 | 24715476 | 24715517 | 42 | |
| Insertion | Chr3 | 34963911 | 34963911 | 1 | LOC_Os03g61690 |
| Insertion | Chr4 | 26319012 | 26319012 | 1 | |
| Insertion | Chr4 | 9567564 | 9567575 | 12 | |
| Insertion | Chr6 | 6623458 | 6623458 | 1 | |
| Insertion | Chr7 | 11810010 | 11810060 | 51 | |
| Insertion | Chr7 | 3555716 | 3555733 | 18 | |
| Insertion | Chr8 | 11214525 | 11214525 | 1 | |
| Insertion | Chr8 | 17189354 | 17189354 | 1 | |
| Insertion | Chr9 | 16695194 | 16695194 | 1 | |
| Insertion | Chr9 | 3239974 | 3239974 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 13001335 | 15255404 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 973000 | Chr5 | 12352448 | LOC_Os05g20990 |
| Translocation | Chr8 | 973009 | Chr5 | 12352213 | LOC_Os05g20990 |
| Translocation | Chr5 | 12352537 | Chr2 | 4262352 | LOC_Os05g20990 |
| Translocation | Chr5 | 12353491 | Chr2 | 4262307 | LOC_Os05g20990 |
| Translocation | Chr10 | 12942940 | Chr2 | 21839457 | LOC_Os10g25110 |
| Translocation | Chr6 | 29650647 | Chr2 | 32946923 | LOC_Os02g53780 |
| Translocation | Chr6 | 29650648 | Chr2 | 32946924 | LOC_Os02g53780 |
