Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2118-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2118-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 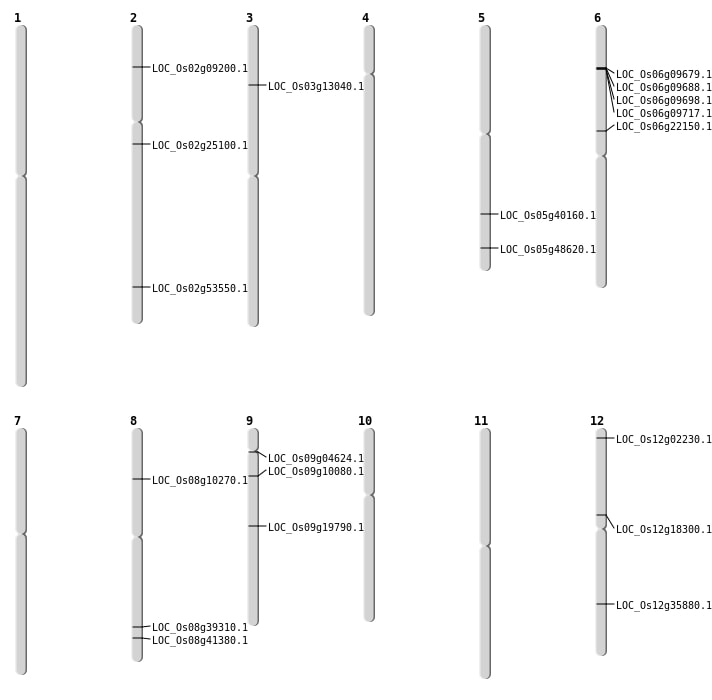
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17833819 | G-T | Homo | ||
| SBS | Chr1 | 20335370 | C-T | Homo | ||
| SBS | Chr1 | 23044397 | T-G | HET | ||
| SBS | Chr1 | 23558315 | G-A | Homo | ||
| SBS | Chr1 | 24149914 | A-G | HET | ||
| SBS | Chr1 | 38653332 | G-T | HET | ||
| SBS | Chr1 | 618276 | C-T | Homo | ||
| SBS | Chr11 | 11477660 | G-A | Homo | ||
| SBS | Chr11 | 2292953 | A-T | HET | ||
| SBS | Chr12 | 10558190 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g18300 |
| SBS | Chr12 | 115558 | G-T | HET | ||
| SBS | Chr12 | 13311053 | C-A | Homo | ||
| SBS | Chr12 | 20728639 | C-T | HET | ||
| SBS | Chr12 | 21864857 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g35880 |
| SBS | Chr2 | 14573277 | C-A | HET | ||
| SBS | Chr2 | 14573278 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g25100 |
| SBS | Chr2 | 15380083 | C-T | HET | ||
| SBS | Chr2 | 29347470 | C-T | Homo | ||
| SBS | Chr2 | 32764606 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g53550 |
| SBS | Chr2 | 33875706 | A-T | HET | ||
| SBS | Chr3 | 1337746 | T-C | Homo | ||
| SBS | Chr3 | 16213515 | G-T | HET | ||
| SBS | Chr3 | 16633641 | G-A | Homo | ||
| SBS | Chr3 | 19948924 | G-A | HET | ||
| SBS | Chr3 | 26082153 | G-T | HET | ||
| SBS | Chr4 | 30007425 | A-T | HET | ||
| SBS | Chr5 | 17707760 | A-G | Homo | ||
| SBS | Chr5 | 19341160 | A-G | Homo | ||
| SBS | Chr5 | 27872508 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g48620 |
| SBS | Chr5 | 2982108 | C-T | Homo | ||
| SBS | Chr6 | 12369862 | C-T | HET | ||
| SBS | Chr6 | 12860247 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g22150 |
| SBS | Chr6 | 16196322 | A-T | HET | ||
| SBS | Chr6 | 17634657 | C-T | HET | ||
| SBS | Chr6 | 21683045 | A-T | HET | ||
| SBS | Chr6 | 5111272 | G-A | Homo | ||
| SBS | Chr8 | 11873867 | A-G | HET | ||
| SBS | Chr9 | 11844025 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g19790 |
| SBS | Chr9 | 11844026 | C-A | HET | ||
| SBS | Chr9 | 1261486 | G-A | Homo | ||
| SBS | Chr9 | 5495450 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g10080 |
Deletions: 9
Insertions: 7
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 693384 | 693803 | LOC_Os12g02230 |
| Inversion | Chr6 | 2912470 | 2912849 | |
| Inversion | Chr11 | 3933441 | 3933620 | |
| Inversion | Chr3 | 7048378 | 7048821 | LOC_Os03g13040 |
| Inversion | Chr6 | 13503543 | 13503902 | |
| Inversion | Chr6 | 21589003 | 21589411 | |
| Inversion | Chr8 | 24841927 | 24842427 | LOC_Os08g39310 |
| Inversion | Chr12 | 25721072 | 25721456 | |
| Inversion | Chr1 | 35487686 | 35488109 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 2459500 | Chr8 | 5973131 | 2 |
| Translocation | Chr8 | 26132308 | Chr2 | 4726018 | |
| Translocation | Chr8 | 26136924 | Chr2 | 4725998 | 2 |
| Translocation | Chr11 | 27797915 | Chr5 | 23589673 | 2 |
