Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2139-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2139-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 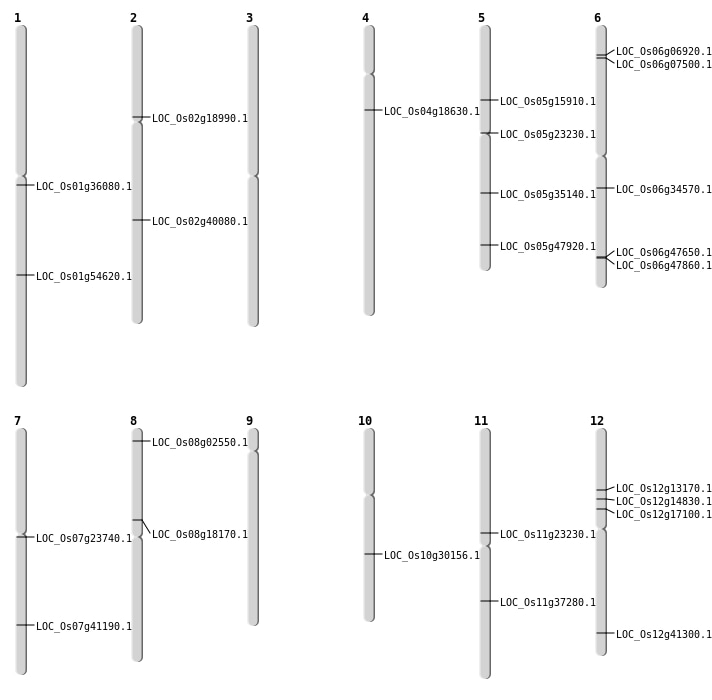
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15341423 | C-G | Homo | ||
| SBS | Chr1 | 25857248 | T-A | Homo | ||
| SBS | Chr1 | 39905634 | A-C | HET | ||
| SBS | Chr1 | 8042139 | G-A | HET | ||
| SBS | Chr10 | 5342253 | A-T | HET | ||
| SBS | Chr10 | 7279848 | T-C | HET | ||
| SBS | Chr11 | 10723497 | C-A | HET | ||
| SBS | Chr11 | 13386059 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g23230 |
| SBS | Chr11 | 13619777 | T-C | HET | ||
| SBS | Chr11 | 16521115 | G-T | HET | ||
| SBS | Chr12 | 10789411 | T-C | Homo | ||
| SBS | Chr12 | 24749238 | C-T | HET | ||
| SBS | Chr12 | 3739095 | C-A | HET | ||
| SBS | Chr12 | 9792546 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g17100 |
| SBS | Chr12 | 9862420 | A-G | Homo | ||
| SBS | Chr2 | 2080935 | C-G | HET | ||
| SBS | Chr2 | 7037074 | G-A | HET | ||
| SBS | Chr3 | 4150496 | G-C | HET | ||
| SBS | Chr4 | 10301197 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18630 |
| SBS | Chr4 | 10396847 | G-T | HET | ||
| SBS | Chr4 | 11148033 | C-T | Homo | ||
| SBS | Chr4 | 1456422 | C-T | HET | ||
| SBS | Chr5 | 27470791 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g47920 |
| SBS | Chr5 | 4838390 | G-A | Homo | ||
| SBS | Chr5 | 5505943 | G-A | HET | ||
| SBS | Chr6 | 22063425 | T-A | HET | ||
| SBS | Chr6 | 28854759 | A-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g47650 |
| SBS | Chr6 | 3608728 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g07500 |
| SBS | Chr6 | 8727930 | C-T | HET | ||
| SBS | Chr8 | 26515270 | G-A | Homo | ||
| SBS | Chr9 | 20772398 | T-C | HET |
Deletions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 2216494 | 2216497 | 3 | |
| Deletion | Chr2 | 4814465 | 4814467 | 2 | |
| Deletion | Chr6 | 6476739 | 6476749 | 10 | |
| Deletion | Chr6 | 8292116 | 8292124 | 8 | |
| Deletion | Chr5 | 10443252 | 10443256 | 4 | |
| Deletion | Chr3 | 13812060 | 13812065 | 5 | |
| Deletion | Chr2 | 21308387 | 21308388 | 1 | |
| Deletion | Chr7 | 24662145 | 24662151 | 6 | LOC_Os07g41190 |
| Deletion | Chr12 | 25613226 | 25613231 | 5 | LOC_Os12g41300 |
| Deletion | Chr7 | 26723919 | 26723922 | 3 | |
| Deletion | Chr6 | 27246999 | 27247001 | 2 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 719531 | 719531 | 1 | |
| Insertion | Chr7 | 7882486 | 7882486 | 1 |
Inversions: 39
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 413576 | 413810 | |
| Inversion | Chr8 | 1053381 | 1053511 | LOC_Os08g02550 |
| Inversion | Chr4 | 2807463 | 2807866 | |
| Inversion | Chr6 | 3283168 | 3283507 | LOC_Os06g06920 |
| Inversion | Chr3 | 3388137 | 3388351 | |
| Inversion | Chr1 | 5780061 | 5780403 | |
| Inversion | Chr2 | 5887583 | 5887936 | |
| Inversion | Chr11 | 7023948 | 7024350 | |
| Inversion | Chr12 | 7334422 | 7334620 | LOC_Os12g13170 |
| Inversion | Chr12 | 8488560 | 8488831 | LOC_Os12g14830 |
| Inversion | Chr5 | 8977207 | 8977359 | LOC_Os05g15910 |
| Inversion | Chr2 | 11094766 | 11095099 | LOC_Os02g18990 |
| Inversion | Chr8 | 11141077 | 11141399 | LOC_Os08g18170 |
| Inversion | Chr3 | 12227376 | 12227480 | |
| Inversion | Chr5 | 13259515 | 13259803 | LOC_Os05g23230 |
| Inversion | Chr4 | 13286304 | 13286454 | |
| Inversion | Chr7 | 13405861 | 13406021 | LOC_Os07g23740 |
| Inversion | Chr2 | 14469614 | 14469846 | |
| Inversion | Chr10 | 15675349 | 15675535 | LOC_Os10g30156 |
| Inversion | Chr6 | 16341052 | 16341468 | |
| Inversion | Chr1 | 17050041 | 17050571 | |
| Inversion | Chr5 | 19452186 | 19452535 | |
| Inversion | Chr1 | 19967236 | 19967341 | LOC_Os01g36080 |
| Inversion | Chr6 | 20109878 | 20110259 | LOC_Os06g34570 |
| Inversion | Chr4 | 20161456 | 20161657 | |
| Inversion | Chr10 | 20393225 | 20393549 | |
| Inversion | Chr5 | 20872979 | 20873376 | LOC_Os05g35140 |
| Inversion | Chr6 | 20928647 | 20928753 | |
| Inversion | Chr4 | 21347590 | 21347698 | |
| Inversion | Chr11 | 22018265 | 22018741 | LOC_Os11g37280 |
| Inversion | Chr1 | 23651694 | 23651818 | |
| Inversion | Chr1 | 23921361 | 23921598 | |
| Inversion | Chr2 | 24268786 | 24269148 | LOC_Os02g40080 |
| Inversion | Chr1 | 26321142 | 26321366 | |
| Inversion | Chr3 | 26621791 | 26622208 | |
| Inversion | Chr4 | 27082655 | 27082889 | |
| Inversion | Chr4 | 28923939 | 28924137 | |
| Inversion | Chr6 | 28963302 | 28963780 | LOC_Os06g47860 |
| Inversion | Chr1 | 31426860 | 31427075 | LOC_Os01g54620 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 11838703 | Chr3 | 2378255 | |
| Translocation | Chr12 | 11839092 | Chr3 | 2378255 | |
| Translocation | Chr8 | 16402728 | Chr5 | 29537150 | |
| Translocation | Chr11 | 18164434 | Chr10 | 19652802 | |
| Translocation | Chr12 | 20178690 | Chr2 | 17732469 | |
| Translocation | Chr11 | 26002459 | Chr6 | 20608561 | |
| Translocation | Chr12 | 27530567 | Chr1 | 21128826 |
