Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2140-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2140-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 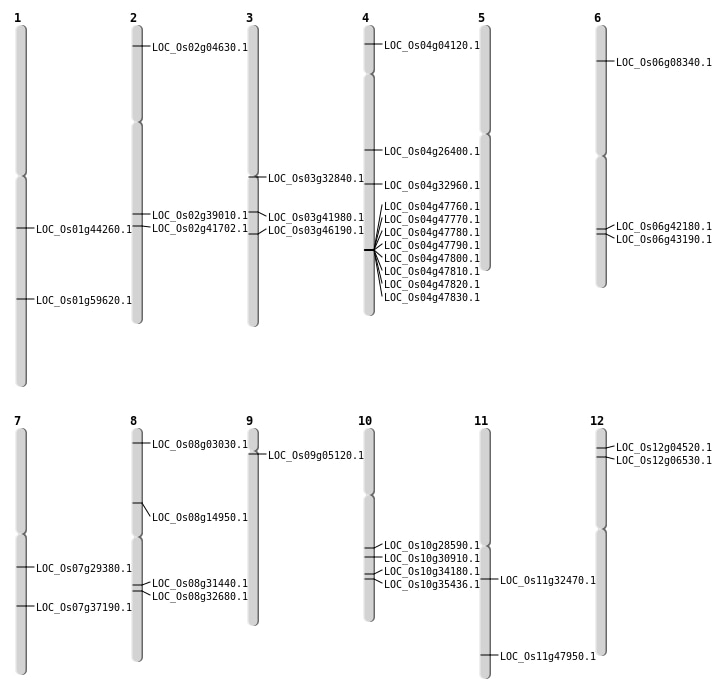
|
| Variant Information |
|---|
Single base substitutions: 53
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14125656 | G-A | HET | ||
| SBS | Chr1 | 32290610 | C-T | Homo | ||
| SBS | Chr1 | 40261481 | C-T | Homo | ||
| SBS | Chr11 | 27826120 | T-A | HET | ||
| SBS | Chr12 | 1926211 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g04520 |
| SBS | Chr12 | 24361341 | T-C | HET | ||
| SBS | Chr12 | 26057012 | G-C | HET | ||
| SBS | Chr12 | 3161144 | A-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g06530 |
| SBS | Chr12 | 7646688 | G-T | HET | ||
| SBS | Chr2 | 20212545 | G-A | HET | ||
| SBS | Chr2 | 2312558 | T-C | Homo | ||
| SBS | Chr2 | 23565278 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g39010 |
| SBS | Chr2 | 25038266 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g41702 |
| SBS | Chr2 | 6125653 | C-T | Homo | ||
| SBS | Chr2 | 7100450 | A-C | HET | ||
| SBS | Chr2 | 9464848 | T-C | HET | ||
| SBS | Chr2 | 9464849 | G-A | HET | ||
| SBS | Chr3 | 14867888 | C-T | Homo | ||
| SBS | Chr3 | 17479849 | T-G | HET | ||
| SBS | Chr3 | 18786272 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g32840 |
| SBS | Chr3 | 18808588 | G-C | HET | ||
| SBS | Chr3 | 19750285 | C-T | HET | ||
| SBS | Chr3 | 19750845 | G-A | HET | ||
| SBS | Chr3 | 22027551 | T-G | HET | ||
| SBS | Chr3 | 23323707 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g41980 |
| SBS | Chr4 | 13190127 | C-T | HET | ||
| SBS | Chr4 | 14674507 | C-T | HET | ||
| SBS | Chr4 | 15397422 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g26400 |
| SBS | Chr4 | 1849019 | G-C | HET | ||
| SBS | Chr4 | 1896971 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g04120 |
| SBS | Chr4 | 1896972 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g04120 |
| SBS | Chr4 | 8524407 | G-A | Homo | ||
| SBS | Chr5 | 15542270 | T-G | HET | ||
| SBS | Chr5 | 22004796 | G-A | HET | ||
| SBS | Chr5 | 22225345 | C-A | HET | ||
| SBS | Chr5 | 27667835 | A-G | HET | ||
| SBS | Chr6 | 1014775 | G-T | Homo | ||
| SBS | Chr6 | 25323014 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g42180 |
| SBS | Chr6 | 29530613 | C-T | HET | ||
| SBS | Chr6 | 30385016 | G-A | HET | ||
| SBS | Chr6 | 6548286 | T-C | HET | ||
| SBS | Chr7 | 1447994 | C-T | HET | ||
| SBS | Chr7 | 17645228 | C-T | HET | ||
| SBS | Chr7 | 21772372 | A-G | HET | ||
| SBS | Chr7 | 22796251 | T-G | HET | ||
| SBS | Chr7 | 26786190 | G-A | HET | ||
| SBS | Chr8 | 1351271 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g03030 |
| SBS | Chr8 | 26258565 | T-C | HET | ||
| SBS | Chr8 | 26258566 | A-T | HET | ||
| SBS | Chr8 | 9017476 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g14950 |
| SBS | Chr9 | 2748151 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g05120 |
| SBS | Chr9 | 4621560 | A-G | HET | ||
| SBS | Chr9 | 47341 | C-T | HET |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 3060030 | 3060043 | 13 | |
| Deletion | Chr6 | 4041133 | 4041137 | 4 | LOC_Os06g08340 |
| Deletion | Chr5 | 6364226 | 6364227 | 1 | |
| Deletion | Chr2 | 7060828 | 7060847 | 19 | |
| Deletion | Chr7 | 7687536 | 7687537 | 1 | |
| Deletion | Chr8 | 7833139 | 7833143 | 4 | |
| Deletion | Chr2 | 8827270 | 8827276 | 6 | |
| Deletion | Chr1 | 10424099 | 10424100 | 1 | |
| Deletion | Chr6 | 15901055 | 15901063 | 8 | |
| Deletion | Chr11 | 16158581 | 16158588 | 7 | |
| Deletion | Chr10 | 18937732 | 18937742 | 10 | LOC_Os10g35436 |
| Deletion | Chr7 | 22284243 | 22284245 | 2 | LOC_Os07g37190 |
| Deletion | Chr6 | 25964901 | 25964907 | 6 | LOC_Os06g43190 |
| Deletion | Chr4 | 28333001 | 28383000 | 49999 | 8 |
| Deletion | Chr1 | 41383221 | 41383242 | 21 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 36298999 | 36299012 | 14 | |
| Insertion | Chr6 | 1193825 | 1193825 | 1 | |
| Insertion | Chr9 | 13061771 | 13061772 | 2 |
Inversions: 21
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 1613429 | 1613635 | |
| Inversion | Chr1 | 2883522 | 2883943 | |
| Inversion | Chr11 | 6260080 | 6260311 | |
| Inversion | Chr10 | 14890261 | 14890386 | LOC_Os10g28590 |
| Inversion | Chr10 | 16121948 | 16122186 | LOC_Os10g30910 |
| Inversion | Chr2 | 16813496 | 16813623 | |
| Inversion | Chr6 | 16887985 | 16888270 | |
| Inversion | Chr7 | 17253933 | 17254577 | LOC_Os07g29380 |
| Inversion | Chr10 | 18243275 | 18243509 | LOC_Os10g34180 |
| Inversion | Chr11 | 19171921 | 19172353 | LOC_Os11g32470 |
| Inversion | Chr8 | 19446439 | 19446697 | LOC_Os08g31440 |
| Inversion | Chr2 | 19487139 | 19487400 | |
| Inversion | Chr4 | 19926583 | 19926889 | LOC_Os04g32960 |
| Inversion | Chr8 | 20244872 | 20245406 | LOC_Os08g32680 |
| Inversion | Chr3 | 22086688 | 22086924 | |
| Inversion | Chr1 | 25383228 | 25383436 | LOC_Os01g44260 |
| Inversion | Chr3 | 26118900 | 26119070 | LOC_Os03g46190 |
| Inversion | Chr8 | 27147021 | 27147167 | |
| Inversion | Chr11 | 28922807 | 28922947 | LOC_Os11g47950 |
| Inversion | Chr4 | 29042615 | 29042878 | |
| Inversion | Chr1 | 34480460 | 34480650 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 3667504 | Chr2 | 2071566 | 2 |
