Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2146-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2146-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 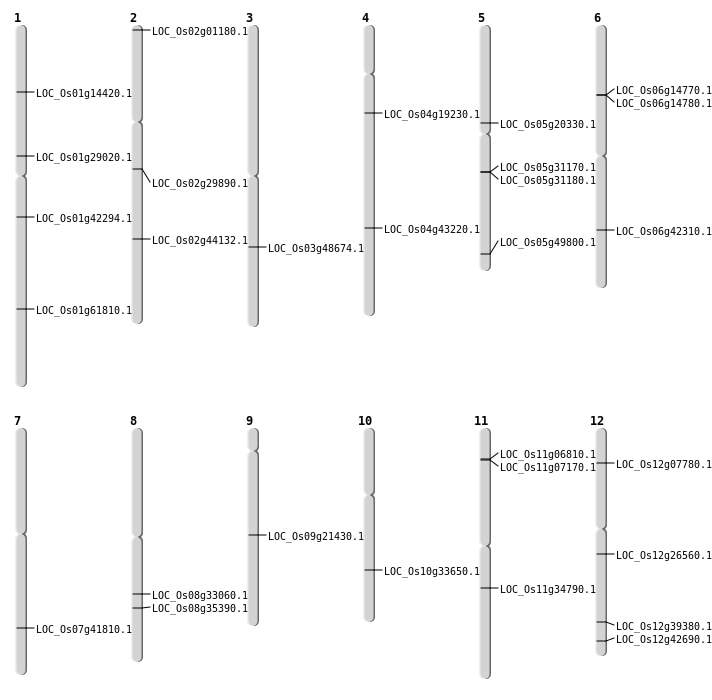
|
| Variant Information |
|---|
Single base substitutions: 62
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12179166 | T-C | HET | ||
| SBS | Chr1 | 16248976 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g29020 |
| SBS | Chr1 | 16358198 | A-G | HET | ||
| SBS | Chr1 | 37074571 | A-G | HET | ||
| SBS | Chr1 | 41780515 | T-C | HET | ||
| SBS | Chr10 | 12611498 | G-A | HET | ||
| SBS | Chr10 | 22955819 | G-A | HET | ||
| SBS | Chr10 | 4490047 | G-C | Homo | ||
| SBS | Chr10 | 7318955 | G-A | HET | ||
| SBS | Chr11 | 10882639 | T-C | HET | ||
| SBS | Chr11 | 15864208 | A-G | HET | ||
| SBS | Chr11 | 22998711 | C-T | HET | ||
| SBS | Chr11 | 3336105 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g06810 |
| SBS | Chr11 | 7552729 | G-C | HET | ||
| SBS | Chr11 | 8726651 | A-G | HET | ||
| SBS | Chr12 | 12091676 | T-A | Homo | ||
| SBS | Chr12 | 15012932 | A-G | HET | ||
| SBS | Chr12 | 15407823 | T-G | HET | ||
| SBS | Chr12 | 24302829 | C-G | HET | ||
| SBS | Chr12 | 26527347 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g42690 |
| SBS | Chr12 | 3466766 | G-A | HET | ||
| SBS | Chr12 | 7401851 | G-A | HET | ||
| SBS | Chr2 | 10749657 | T-A | HET | ||
| SBS | Chr2 | 17784504 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g29890 |
| SBS | Chr2 | 19702646 | A-T | HET | ||
| SBS | Chr2 | 19702647 | A-T | HET | ||
| SBS | Chr2 | 844785 | A-G | HET | ||
| SBS | Chr3 | 19300587 | G-A | Homo | ||
| SBS | Chr3 | 31310747 | G-A | HET | ||
| SBS | Chr3 | 4649508 | T-C | HET | ||
| SBS | Chr3 | 4649509 | C-A | HET | ||
| SBS | Chr3 | 5139591 | T-C | HET | ||
| SBS | Chr4 | 10680566 | T-C | HET | SPLICE_SITE_DONOR | LOC_Os04g19230 |
| SBS | Chr4 | 13160012 | A-T | HET | ||
| SBS | Chr4 | 13160013 | G-T | HET | ||
| SBS | Chr4 | 13258873 | A-T | HET | ||
| SBS | Chr4 | 23218693 | G-A | HET | ||
| SBS | Chr4 | 25577574 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g43220 |
| SBS | Chr4 | 28252042 | A-G | HET | ||
| SBS | Chr4 | 34362475 | A-G | HET | ||
| SBS | Chr5 | 11924424 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g20330 |
| SBS | Chr5 | 12893697 | T-A | Homo | ||
| SBS | Chr5 | 20830494 | C-G | HET | ||
| SBS | Chr5 | 21899104 | T-A | HET | ||
| SBS | Chr5 | 22662325 | T-C | HET | ||
| SBS | Chr5 | 8301792 | A-G | HET | ||
| SBS | Chr6 | 26499374 | C-G | HET | ||
| SBS | Chr6 | 27602013 | G-A | HET | ||
| SBS | Chr6 | 9180037 | C-T | HET | ||
| SBS | Chr7 | 23902626 | A-G | HET | ||
| SBS | Chr7 | 7439979 | G-C | HET | ||
| SBS | Chr8 | 15303737 | T-C | Homo | ||
| SBS | Chr8 | 15504348 | C-G | HET | ||
| SBS | Chr8 | 1965597 | T-A | Homo | ||
| SBS | Chr8 | 22312356 | C-A | Homo | STOP_GAINED | LOC_Os08g35390 |
| SBS | Chr8 | 3627515 | G-A | HET | ||
| SBS | Chr8 | 8165254 | A-G | HET | ||
| SBS | Chr9 | 11597354 | G-A | HET | ||
| SBS | Chr9 | 14765291 | G-A | HET | ||
| SBS | Chr9 | 17900424 | T-A | HET | ||
| SBS | Chr9 | 7026274 | G-A | HET | ||
| SBS | Chr9 | 9144410 | A-C | HET |
Deletions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 5242413 | 5242414 | 1 | |
| Deletion | Chr6 | 8339001 | 8375000 | 35999 | 2 |
| Deletion | Chr6 | 8650972 | 8650973 | 1 | |
| Deletion | Chr4 | 10140149 | 10140151 | 2 | |
| Deletion | Chr8 | 10654288 | 10654298 | 10 | |
| Deletion | Chr3 | 11170703 | 11170713 | 10 | |
| Deletion | Chr12 | 15524502 | 15524507 | 5 | LOC_Os12g26560 |
| Deletion | Chr5 | 18137001 | 18147000 | 9999 | 2 |
| Deletion | Chr7 | 18484230 | 18484235 | 5 | |
| Deletion | Chr10 | 19894223 | 19894242 | 19 | |
| Deletion | Chr6 | 24100540 | 24100542 | 2 | |
| Deletion | Chr3 | 27747939 | 27747942 | 3 | LOC_Os03g48674 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 17243800 | 17243800 | 1 |
Inversions: 30
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 102688 | 102988 | LOC_Os02g01180 |
| Inversion | Chr12 | 871961 | 872314 | |
| Inversion | Chr4 | 3473910 | 3474241 | |
| Inversion | Chr11 | 3583513 | 3583919 | LOC_Os11g07170 |
| Inversion | Chr12 | 3933455 | 3933903 | LOC_Os12g07780 |
| Inversion | Chr1 | 4596820 | 4597115 | |
| Inversion | Chr8 | 4747227 | 4747345 | |
| Inversion | Chr4 | 5047530 | 5047751 | |
| Inversion | Chr2 | 5215697 | 5216066 | |
| Inversion | Chr8 | 5530609 | 5530928 | |
| Inversion | Chr1 | 8068862 | 8069499 | LOC_Os01g14420 |
| Inversion | Chr10 | 12148648 | 12149002 | |
| Inversion | Chr1 | 16435112 | 16435415 | |
| Inversion | Chr10 | 17743988 | 17744121 | LOC_Os10g33650 |
| Inversion | Chr1 | 20236288 | 20236589 | |
| Inversion | Chr12 | 20244783 | 20245036 | |
| Inversion | Chr11 | 20382417 | 20382833 | LOC_Os11g34790 |
| Inversion | Chr8 | 20431347 | 20540778 | 2 |
| Inversion | Chr12 | 21475365 | 21475531 | |
| Inversion | Chr3 | 22280343 | 22280539 | |
| Inversion | Chr1 | 23980896 | 23981338 | LOC_Os01g42294 |
| Inversion | Chr12 | 24220544 | 24220690 | LOC_Os12g39380 |
| Inversion | Chr7 | 25045288 | 25045591 | LOC_Os07g41810 |
| Inversion | Chr6 | 25417762 | 25417983 | LOC_Os06g42310 |
| Inversion | Chr2 | 26699568 | 26699846 | LOC_Os02g44132 |
| Inversion | Chr5 | 27021023 | 27021306 | |
| Inversion | Chr3 | 27941132 | 27941452 | |
| Inversion | Chr5 | 28582234 | 28582383 | LOC_Os05g49800 |
| Inversion | Chr1 | 33699897 | 33700096 | |
| Inversion | Chr1 | 35758214 | 35758411 | LOC_Os01g61810 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 4497852 | Chr5 | 665832 | |
| Translocation | Chr10 | 8390288 | Chr6 | 8374281 | 2 |
| Translocation | Chr9 | 12955649 | Chr6 | 8210715 | LOC_Os09g21430 |
