Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2151-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2151-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 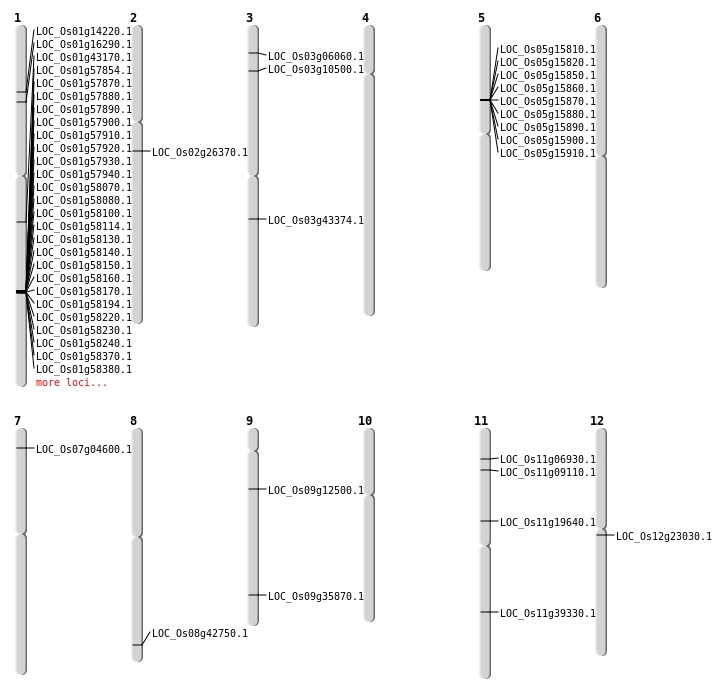
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10269876 | G-T | HET | ||
| SBS | Chr1 | 12991637 | T-C | HET | ||
| SBS | Chr1 | 29226278 | G-A | Homo | ||
| SBS | Chr1 | 5404845 | G-A | HET | ||
| SBS | Chr1 | 7072879 | A-G | HET | ||
| SBS | Chr1 | 9389035 | T-A | HET | ||
| SBS | Chr11 | 11318811 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g19640 |
| SBS | Chr11 | 23417138 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g39330 |
| SBS | Chr12 | 13017811 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g23030 |
| SBS | Chr12 | 21501132 | A-G | Homo | ||
| SBS | Chr2 | 19266218 | G-T | HET | ||
| SBS | Chr2 | 2330976 | T-C | HET | ||
| SBS | Chr3 | 3037308 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g06060 |
| SBS | Chr3 | 35530334 | T-C | HET | ||
| SBS | Chr4 | 11339248 | G-T | Homo | ||
| SBS | Chr4 | 17266156 | G-A | HET | ||
| SBS | Chr4 | 19654164 | G-A | HET | ||
| SBS | Chr4 | 28355787 | T-C | HET | ||
| SBS | Chr4 | 7422407 | G-C | HET | ||
| SBS | Chr4 | 8314915 | G-T | HET | ||
| SBS | Chr5 | 12412226 | G-A | HET | ||
| SBS | Chr5 | 21066958 | G-A | Homo | ||
| SBS | Chr5 | 6500108 | G-A | HET | ||
| SBS | Chr6 | 14577673 | C-T | HET | ||
| SBS | Chr6 | 19905567 | C-A | HET | ||
| SBS | Chr6 | 19905570 | C-T | HET | ||
| SBS | Chr6 | 8552483 | G-A | HET | ||
| SBS | Chr8 | 19649891 | A-G | HET | ||
| SBS | Chr8 | 21353429 | C-T | HET | ||
| SBS | Chr9 | 21268279 | G-A | HET | ||
| SBS | Chr9 | 3772425 | T-C | HET | ||
| SBS | Chr9 | 4955326 | C-T | HET | ||
| SBS | Chr9 | 7168502 | A-T | HET |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 668968 | 668975 | 7 | |
| Deletion | Chr2 | 2050296 | 2050306 | 10 | |
| Deletion | Chr9 | 3029982 | 3029984 | 2 | |
| Deletion | Chr4 | 3136741 | 3136749 | 8 | |
| Deletion | Chr5 | 8922001 | 8976000 | 53999 | 9 |
| Deletion | Chr1 | 14945742 | 14945744 | 2 | |
| Deletion | Chr6 | 17298043 | 17298051 | 8 | |
| Deletion | Chr6 | 22660294 | 22660306 | 12 | |
| Deletion | Chr3 | 22865538 | 22865539 | 1 | |
| Deletion | Chr4 | 27410360 | 27410369 | 9 | |
| Deletion | Chr4 | 32587618 | 32587620 | 2 | |
| Deletion | Chr1 | 33458001 | 33495000 | 36999 | 9 |
| Deletion | Chr1 | 33591001 | 33665000 | 73999 | 13 |
| Deletion | Chr1 | 33726001 | 33810000 | 83999 | 13 |
| Deletion | Chr1 | 36644452 | 36644453 | 1 | LOC_Os01g63230 |
| Deletion | Chr1 | 36678474 | 36678494 | 20 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 9519492 | 9519492 | 1 | |
| Insertion | Chr5 | 5851224 | 5851224 | 1 |
Inversions: 25
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 1502773 | 1502874 | |
| Inversion | Chr7 | 2048955 | 2049507 | LOC_Os07g04600 |
| Inversion | Chr2 | 2515622 | 2515912 | |
| Inversion | Chr11 | 4851295 | 4851510 | LOC_Os11g09110 |
| Inversion | Chr11 | 5089898 | 5090371 | |
| Inversion | Chr3 | 5343197 | 5343396 | LOC_Os03g10500 |
| Inversion | Chr9 | 6984913 | 6985187 | |
| Inversion | Chr10 | 7330552 | 7330808 | |
| Inversion | Chr1 | 7973068 | 7973427 | LOC_Os01g14220 |
| Inversion | Chr12 | 8499695 | 8499891 | |
| Inversion | Chr12 | 8499754 | 8499967 | |
| Inversion | Chr12 | 8499810 | 8500010 | |
| Inversion | Chr1 | 8847242 | 9236989 | 2 |
| Inversion | Chr5 | 11245242 | 11245751 | |
| Inversion | Chr1 | 12947056 | 12947194 | |
| Inversion | Chr11 | 13371269 | 13371531 | |
| Inversion | Chr9 | 15370592 | 15371049 | |
| Inversion | Chr2 | 15482579 | 15482761 | LOC_Os02g26370 |
| Inversion | Chr5 | 16350561 | 16351095 | |
| Inversion | Chr9 | 20642892 | 20643155 | LOC_Os09g35870 |
| Inversion | Chr3 | 24185028 | 24185195 | LOC_Os03g43374 |
| Inversion | Chr1 | 24634938 | 24635092 | LOC_Os01g43170 |
| Inversion | Chr4 | 25093509 | 25093785 | |
| Inversion | Chr8 | 27056879 | 27056982 | LOC_Os08g42750 |
| Inversion | Chr1 | 29669038 | 29669265 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 1877733 | Chr2 | 13467040 | |
| Translocation | Chr11 | 3421472 | Chr9 | 7162016 | LOC_Os11g06930 |
| Translocation | Chr11 | 3421485 | Chr9 | 7169343 | 2 |
| Translocation | Chr2 | 18067236 | Chr1 | 8221492 |
