Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2153-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2153-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 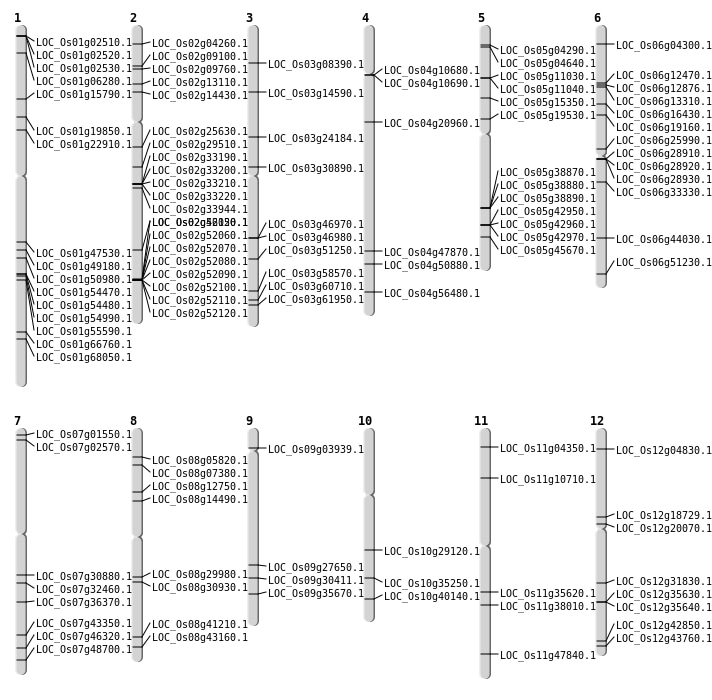
|
| Variant Information |
|---|
Single base substitutions: 23
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 1093802 | G-A | HET | ||
| SBS | Chr11 | 5942146 | T-C | HET | ||
| SBS | Chr12 | 10397866 | G-A | HET | ||
| SBS | Chr12 | 11674209 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g20070 |
| SBS | Chr12 | 1395276 | A-T | HET | ||
| SBS | Chr12 | 8486405 | T-C | HET | ||
| SBS | Chr12 | 8769411 | G-A | Homo | ||
| SBS | Chr3 | 35107250 | C-A | Homo | ||
| SBS | Chr3 | 5274250 | T-C | Homo | ||
| SBS | Chr4 | 296765 | T-A | HET | ||
| SBS | Chr5 | 1939728 | T-A | HET | ||
| SBS | Chr5 | 28932231 | A-G | HET | ||
| SBS | Chr6 | 28131355 | C-G | HET | ||
| SBS | Chr7 | 10727567 | T-G | Homo | ||
| SBS | Chr7 | 18984209 | A-T | HET | ||
| SBS | Chr7 | 19311589 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g32460 |
| SBS | Chr7 | 21749138 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36370 |
| SBS | Chr7 | 22342272 | G-T | HET | ||
| SBS | Chr7 | 27073489 | A-G | HET | ||
| SBS | Chr8 | 5837021 | G-A | HET | ||
| SBS | Chr8 | 8300715 | T-C | HET | ||
| SBS | Chr8 | 9406953 | G-A | Homo | ||
| SBS | Chr9 | 16410731 | A-T | HET |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 67897 | 67912 | 15 | |
| Deletion | Chr1 | 814001 | 830000 | 15999 | 3 |
| Deletion | Chr5 | 1947464 | 1947465 | 1 | LOC_Os05g04290 |
| Deletion | Chr2 | 5088069 | 5088072 | 3 | |
| Deletion | Chr5 | 6180001 | 6190000 | 9999 | 2 |
| Deletion | Chr6 | 6706942 | 6706944 | 2 | |
| Deletion | Chr10 | 8646076 | 8646077 | 1 | |
| Deletion | Chr8 | 15171857 | 15171948 | 91 | |
| Deletion | Chr10 | 15171891 | 15172212 | 321 | LOC_Os10g29120 |
| Deletion | Chr6 | 16490001 | 16502000 | 11999 | 3 |
| Deletion | Chr11 | 17871439 | 17871807 | 368 | |
| Deletion | Chr3 | 18162770 | 18162776 | 6 | |
| Deletion | Chr2 | 19735001 | 19750000 | 14999 | 4 |
| Deletion | Chr5 | 22801001 | 22814000 | 12999 | 3 |
| Deletion | Chr5 | 24935001 | 24953000 | 17999 | 3 |
| Deletion | Chr5 | 25899944 | 25899946 | 2 | |
| Deletion | Chr3 | 26557001 | 26572000 | 14999 | 2 |
| Deletion | Chr12 | 27174001 | 27188000 | 13999 | LOC_Os12g43760 |
| Deletion | Chr3 | 27609876 | 27609877 | 1 | |
| Deletion | Chr4 | 30207893 | 30207899 | 6 | |
| Deletion | Chr2 | 31857001 | 31901000 | 43999 | 8 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 8280328 | 8280328 | 1 | |
| Insertion | Chr3 | 19556472 | 19556472 | 1 | |
| Insertion | Chr4 | 2207625 | 2207625 | 1 | |
| Insertion | Chr4 | 7914316 | 7914319 | 4 | |
| Insertion | Chr7 | 18280214 | 18280214 | 1 | LOC_Os07g30880 |
| Insertion | Chr8 | 7532153 | 7532153 | 1 |
Inversions: 149
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 46504 | 46801 | |
| Inversion | Chr7 | 343961 | 344415 | LOC_Os07g01550 |
| Inversion | Chr1 | 528607 | 528773 | |
| Inversion | Chr3 | 850465 | 850574 | |
| Inversion | Chr7 | 925779 | 926202 | LOC_Os07g02570 |
| Inversion | Chr2 | 1026956 | 1027200 | |
| Inversion | Chr11 | 1808300 | 1808796 | LOC_Os11g04350 |
| Inversion | Chr6 | 1824480 | 1824729 | LOC_Os06g04300 |
| Inversion | Chr2 | 1881957 | 1882231 | LOC_Os02g04260 |
| Inversion | Chr9 | 2020487 | 2020604 | LOC_Os09g03939 |
| Inversion | Chr5 | 2183474 | 2183857 | LOC_Os05g04640 |
| Inversion | Chr5 | 2208886 | 2328772 | |
| Inversion | Chr5 | 2208893 | 2328780 | |
| Inversion | Chr1 | 2985282 | 2985544 | LOC_Os01g06280 |
| Inversion | Chr8 | 3123566 | 3123674 | LOC_Os08g05820 |
| Inversion | Chr10 | 3592242 | 3592364 | |
| Inversion | Chr7 | 3997075 | 3997568 | |
| Inversion | Chr8 | 4135360 | 4135519 | LOC_Os08g07380 |
| Inversion | Chr3 | 4290756 | 4290983 | LOC_Os03g08390 |
| Inversion | Chr12 | 4678278 | 4678460 | |
| Inversion | Chr2 | 5033771 | 5033965 | LOC_Os02g09760 |
| Inversion | Chr12 | 5488646 | 5489168 | |
| Inversion | Chr4 | 5810957 | 5811372 | 2 |
| Inversion | Chr11 | 5876712 | 5877002 | LOC_Os11g10710 |
| Inversion | Chr3 | 6017935 | 6018047 | |
| Inversion | Chr1 | 6156810 | 6156910 | |
| Inversion | Chr6 | 6767739 | 6767957 | LOC_Os06g12470 |
| Inversion | Chr7 | 6788719 | 6788928 | |
| Inversion | Chr2 | 6971440 | 6971607 | LOC_Os02g13110 |
| Inversion | Chr6 | 7048624 | 7048989 | LOC_Os06g12876 |
| Inversion | Chr11 | 7051283 | 7051484 | |
| Inversion | Chr8 | 7182630 | 7182859 | |
| Inversion | Chr11 | 7202102 | 7202553 | |
| Inversion | Chr6 | 7227135 | 7227314 | |
| Inversion | Chr6 | 7241604 | 7241891 | |
| Inversion | Chr4 | 7284511 | 7284634 | |
| Inversion | Chr6 | 7320139 | 7320298 | LOC_Os06g13310 |
| Inversion | Chr8 | 7553790 | 7554099 | LOC_Os08g12750 |
| Inversion | Chr6 | 7707820 | 7708561 | |
| Inversion | Chr3 | 7915854 | 7916057 | LOC_Os03g14590 |
| Inversion | Chr2 | 7938856 | 7939272 | LOC_Os02g14430 |
| Inversion | Chr5 | 8241582 | 8241733 | |
| Inversion | Chr5 | 8689009 | 8689354 | LOC_Os05g15350 |
| Inversion | Chr8 | 8704270 | 8704534 | LOC_Os08g14490 |
| Inversion | Chr1 | 8894981 | 8895142 | LOC_Os01g15790 |
| Inversion | Chr5 | 9133951 | 9134213 | |
| Inversion | Chr6 | 9393175 | 9393519 | |
| Inversion | Chr6 | 9407500 | 9407679 | LOC_Os06g16430 |
| Inversion | Chr10 | 9508969 | 9509113 | |
| Inversion | Chr12 | 10830706 | 10830904 | LOC_Os12g18729 |
| Inversion | Chr12 | 10830743 | 10831044 | LOC_Os12g18729 |
| Inversion | Chr6 | 10905543 | 10905687 | LOC_Os06g19160 |
| Inversion | Chr1 | 11259647 | 11260116 | LOC_Os01g19850 |
| Inversion | Chr11 | 11623409 | 11623513 | |
| Inversion | Chr5 | 11699464 | 11699581 | |
| Inversion | Chr4 | 11778708 | 11778939 | LOC_Os04g20960 |
| Inversion | Chr6 | 12282014 | 12282437 | |
| Inversion | Chr10 | 12729235 | 12729534 | |
| Inversion | Chr9 | 12824129 | 12824302 | |
| Inversion | Chr1 | 12878712 | 12879014 | LOC_Os01g22910 |
| Inversion | Chr2 | 13253388 | 13253577 | |
| Inversion | Chr3 | 13600486 | 13600838 | |
| Inversion | Chr3 | 13744253 | 13744622 | LOC_Os03g24184 |
| Inversion | Chr7 | 14202703 | 14202979 | |
| Inversion | Chr1 | 14741890 | 14742314 | |
| Inversion | Chr2 | 14984203 | 14984638 | LOC_Os02g25630 |
| Inversion | Chr5 | 15005425 | 15005711 | |
| Inversion | Chr6 | 15218626 | 15218819 | LOC_Os06g25990 |
| Inversion | Chr5 | 15572404 | 15572823 | |
| Inversion | Chr11 | 15610628 | 15610955 | |
| Inversion | Chr7 | 15990102 | 15990259 | |
| Inversion | Chr11 | 16229749 | 16229967 | |
| Inversion | Chr9 | 16825369 | 16825531 | LOC_Os09g27650 |
| Inversion | Chr8 | 16859105 | 16859291 | |
| Inversion | Chr5 | 16952047 | 16952169 | |
| Inversion | Chr11 | 17214024 | 17214462 | |
| Inversion | Chr3 | 17269204 | 17269563 | |
| Inversion | Chr4 | 17392638 | 17392794 | |
| Inversion | Chr6 | 17401359 | 17401479 | |
| Inversion | Chr2 | 17538267 | 17538473 | LOC_Os02g29510 |
| Inversion | Chr6 | 17542929 | 17543453 | |
| Inversion | Chr3 | 17600550 | 17600793 | LOC_Os03g30890 |
| Inversion | Chr12 | 18266512 | 18266816 | |
| Inversion | Chr8 | 18416111 | 18416263 | LOC_Os08g29980 |
| Inversion | Chr9 | 18513493 | 18513699 | LOC_Os09g30411 |
| Inversion | Chr10 | 18839728 | 18839864 | LOC_Os10g35250 |
| Inversion | Chr8 | 19095993 | 19096295 | LOC_Os08g30930 |
| Inversion | Chr12 | 19151691 | 19151851 | LOC_Os12g31830 |
| Inversion | Chr6 | 19415184 | 19415356 | LOC_Os06g33330 |
| Inversion | Chr9 | 19422591 | 19422919 | |
| Inversion | Chr4 | 19588902 | 19589372 | |
| Inversion | Chr12 | 19898943 | 19899348 | |
| Inversion | Chr2 | 20234623 | 20234811 | LOC_Os02g33944 |
| Inversion | Chr4 | 20493619 | 20494611 | |
| Inversion | Chr9 | 20518699 | 20519280 | 2 |
| Inversion | Chr11 | 20887077 | 20887227 | LOC_Os11g35620 |
| Inversion | Chr10 | 21488597 | 21489237 | LOC_Os10g40140 |
| Inversion | Chr12 | 21666835 | 21667023 | 2 |
| Inversion | Chr10 | 22527594 | 22527734 | |
| Inversion | Chr12 | 22534359 | 22534657 | LOC_Os12g36770 |
| Inversion | Chr11 | 22553503 | 22553876 | LOC_Os11g38010 |
| Inversion | Chr3 | 22612190 | 22612522 | |
| Inversion | Chr10 | 22996687 | 22996970 | |
| Inversion | Chr4 | 23486755 | 23487165 | |
| Inversion | Chr12 | 23523213 | 23523356 | |
| Inversion | Chr3 | 23532712 | 23532903 | |
| Inversion | Chr2 | 24054550 | 24054666 | |
| Inversion | Chr3 | 24651251 | 24651399 | |
| Inversion | Chr12 | 24914383 | 24914582 | |
| Inversion | Chr8 | 25106324 | 25106627 | |
| Inversion | Chr8 | 25364205 | 25365305 | |
| Inversion | Chr12 | 25639773 | 25640189 | |
| Inversion | Chr7 | 25946977 | 25947361 | LOC_Os07g43350 |
| Inversion | Chr8 | 26025012 | 26025224 | LOC_Os08g41210 |
| Inversion | Chr11 | 26131579 | 26131800 | |
| Inversion | Chr5 | 26467492 | 26468052 | 2 |
| Inversion | Chr6 | 26532101 | 26532215 | LOC_Os06g44030 |
| Inversion | Chr12 | 26624652 | 26624790 | LOC_Os12g42850 |
| Inversion | Chr1 | 26640530 | 26640671 | |
| Inversion | Chr1 | 27172251 | 27172470 | LOC_Os01g47530 |
| Inversion | Chr8 | 27297997 | 27298196 | LOC_Os08g43160 |
| Inversion | Chr7 | 27629878 | 27630231 | LOC_Os07g46320 |
| Inversion | Chr2 | 28111696 | 28112296 | LOC_Os02g46120 |
| Inversion | Chr1 | 28272126 | 28272485 | LOC_Os01g49180 |
| Inversion | Chr4 | 28432889 | 28433072 | LOC_Os04g47870 |
| Inversion | Chr1 | 28834064 | 28834206 | |
| Inversion | Chr11 | 28855137 | 28855480 | LOC_Os11g47840 |
| Inversion | Chr7 | 29160286 | 29160449 | LOC_Os07g48700 |
| Inversion | Chr1 | 29290832 | 29291046 | LOC_Os01g50980 |
| Inversion | Chr3 | 29319884 | 29320027 | LOC_Os03g51250 |
| Inversion | Chr4 | 30107861 | 30108210 | LOC_Os04g50880 |
| Inversion | Chr1 | 30638620 | 30638984 | |
| Inversion | Chr6 | 31003723 | 31003918 | LOC_Os06g51230 |
| Inversion | Chr6 | 31077843 | 31078174 | |
| Inversion | Chr1 | 31333469 | 31333749 | 2 |
| Inversion | Chr1 | 31623181 | 31623467 | LOC_Os01g54990 |
| Inversion | Chr1 | 32030734 | 32030847 | LOC_Os01g55590 |
| Inversion | Chr3 | 33349641 | 33350080 | LOC_Os03g58570 |
| Inversion | Chr4 | 33670461 | 33670699 | LOC_Os04g56480 |
| Inversion | Chr2 | 33795829 | 33796006 | |
| Inversion | Chr3 | 34504405 | 34504570 | LOC_Os03g60710 |
| Inversion | Chr3 | 35118309 | 35118531 | LOC_Os03g61950 |
| Inversion | Chr1 | 35182523 | 35182972 | |
| Inversion | Chr1 | 35420523 | 35420775 | |
| Inversion | Chr3 | 36368850 | 36369069 | |
| Inversion | Chr1 | 38770917 | 38771019 | LOC_Os01g66760 |
| Inversion | Chr1 | 39563567 | 39563839 | LOC_Os01g68050 |
| Inversion | Chr1 | 42016252 | 42016357 | |
| Inversion | Chr1 | 42488814 | 42488919 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2071497 | Chr5 | 11351597 | 2 |
| Translocation | Chr6 | 8871880 | Chr1 | 18619979 | |
| Translocation | Chr10 | 9894520 | Chr1 | 23525410 | |
| Translocation | Chr12 | 13468561 | Chr2 | 4662086 | 2 |
| Translocation | Chr12 | 13468842 | Chr2 | 4662088 | 2 |
| Translocation | Chr12 | 19763492 | Chr5 | 28571090 | |
| Translocation | Chr7 | 27046689 | Chr3 | 24872885 |
