Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2243-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2243-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 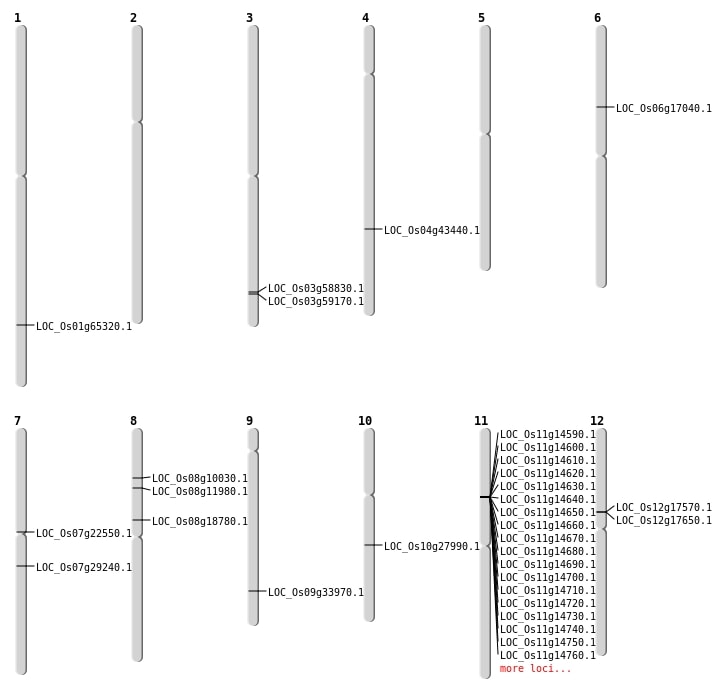
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20184106 | G-A | HET | ||
| SBS | Chr1 | 31160941 | C-T | Homo | ||
| SBS | Chr1 | 37893524 | G-A | HET | STOP_GAINED | LOC_Os01g65320 |
| SBS | Chr1 | 5189564 | C-A | HET | ||
| SBS | Chr1 | 6418307 | G-A | Homo | ||
| SBS | Chr10 | 12697557 | C-G | Homo | ||
| SBS | Chr10 | 1819338 | G-A | HET | ||
| SBS | Chr10 | 2887125 | T-G | HET | ||
| SBS | Chr11 | 21146044 | G-A | HET | ||
| SBS | Chr12 | 19720523 | T-C | HET | ||
| SBS | Chr2 | 16697007 | C-A | HET | ||
| SBS | Chr2 | 20547996 | G-A | HET | ||
| SBS | Chr2 | 20547997 | G-A | HET | ||
| SBS | Chr2 | 20548001 | T-A | HET | ||
| SBS | Chr2 | 20599652 | G-A | HET | ||
| SBS | Chr2 | 26765857 | G-A | HET | ||
| SBS | Chr3 | 32949553 | A-G | HET | ||
| SBS | Chr3 | 33197966 | C-G | HET | ||
| SBS | Chr3 | 4895715 | T-C | HET | ||
| SBS | Chr3 | 8004632 | A-G | HET | ||
| SBS | Chr3 | 9233155 | G-A | HET | ||
| SBS | Chr4 | 10676258 | A-C | Homo | ||
| SBS | Chr4 | 19868053 | G-C | HET | ||
| SBS | Chr4 | 21749586 | G-A | HET | ||
| SBS | Chr4 | 25712365 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g43440 |
| SBS | Chr4 | 7057064 | G-T | HET | ||
| SBS | Chr6 | 15728610 | G-A | HET | ||
| SBS | Chr6 | 9862661 | C-T | HET | ||
| SBS | Chr6 | 9880558 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g17040 |
| SBS | Chr7 | 12688020 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g22550 |
| SBS | Chr7 | 14995053 | T-A | HET | ||
| SBS | Chr7 | 3744454 | C-T | HET | ||
| SBS | Chr8 | 5812806 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g10030 |
| SBS | Chr8 | 7031520 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g11980 |
| SBS | Chr9 | 20055852 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g33970 |
| SBS | Chr9 | 20802058 | G-T | HET | ||
| SBS | Chr9 | 8759909 | G-T | HET |
Deletions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 614294 | 614296 | 2 | |
| Deletion | Chr11 | 8210001 | 8376000 | 165999 | 29 |
| Deletion | Chr11 | 8416001 | 8557000 | 140999 | 22 |
| Deletion | Chr11 | 8700001 | 8775000 | 74999 | 11 |
| Deletion | Chr8 | 11209419 | 11210414 | 995 | LOC_Os08g18780 |
| Deletion | Chr3 | 13202423 | 13202434 | 11 | |
| Deletion | Chr10 | 14502832 | 14502833 | 1 | LOC_Os10g27990 |
| Deletion | Chr6 | 17335168 | 17335174 | 6 | |
| Deletion | Chr10 | 18976757 | 18976760 | 3 | |
| Deletion | Chr5 | 20427842 | 20427848 | 6 | |
| Deletion | Chr6 | 23696924 | 23696925 | 1 | |
| Deletion | Chr3 | 25794334 | 25794335 | 1 | |
| Deletion | Chr7 | 26828531 | 26828532 | 1 | |
| Deletion | Chr2 | 30606206 | 30606207 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 15780243 | 15780244 | 2 | |
| Insertion | Chr5 | 5150992 | 5150995 | 4 | |
| Insertion | Chr6 | 21204836 | 21204836 | 1 | |
| Insertion | Chr6 | 21857133 | 21857133 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 10071497 | 10108813 | 2 |
| Inversion | Chr12 | 10071569 | 10108807 | 2 |
| Inversion | Chr3 | 33502319 | 33694522 | 2 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 1207 | Chr2 | 151 | |
| Translocation | Chr9 | 3001013 | Chr3 | 16253362 | |
| Translocation | Chr4 | 6910503 | Chr3 | 33694526 | 2 |
| Translocation | Chr4 | 6931538 | Chr3 | 33502323 | 2 |
| Translocation | Chr11 | 8777367 | Chr3 | 16253767 | |
| Translocation | Chr12 | 13547459 | Chr11 | 22973797 | 2 |
| Translocation | Chr7 | 17141775 | Chr6 | 16283303 | LOC_Os07g29240 |
