Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2265-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2265-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 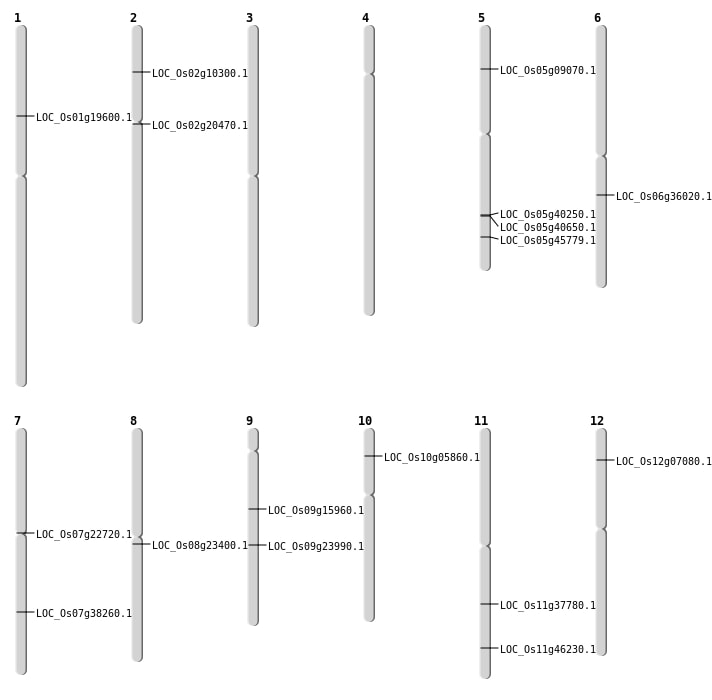
|
| Variant Information |
|---|
Single base substitutions: 48
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10077085 | G-A | HET | ||
| SBS | Chr1 | 10987156 | G-A | Homo | ||
| SBS | Chr1 | 16940253 | G-A | HET | ||
| SBS | Chr1 | 17116821 | G-A | Homo | ||
| SBS | Chr1 | 17116823 | T-C | Homo | ||
| SBS | Chr1 | 27175618 | C-A | HET | ||
| SBS | Chr10 | 16735927 | G-A | HET | ||
| SBS | Chr10 | 66235 | C-A | HET | ||
| SBS | Chr11 | 15689402 | A-C | HET | ||
| SBS | Chr11 | 18885269 | A-G | HET | ||
| SBS | Chr11 | 25557338 | C-T | HET | ||
| SBS | Chr11 | 8216108 | T-C | HET | ||
| SBS | Chr12 | 20995856 | C-A | HET | ||
| SBS | Chr12 | 3470554 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g07080 |
| SBS | Chr2 | 26164771 | C-T | HET | ||
| SBS | Chr2 | 26791662 | A-T | HET | ||
| SBS | Chr3 | 15802928 | G-A | HET | ||
| SBS | Chr3 | 30312317 | C-T | HET | ||
| SBS | Chr3 | 499610 | G-T | HET | ||
| SBS | Chr4 | 10667075 | C-T | HET | ||
| SBS | Chr4 | 15073767 | T-C | HET | ||
| SBS | Chr4 | 15476738 | C-T | HET | ||
| SBS | Chr4 | 16662340 | T-A | HET | ||
| SBS | Chr4 | 27669166 | C-T | HET | ||
| SBS | Chr5 | 14171272 | G-A | HET | ||
| SBS | Chr5 | 18093556 | G-T | HET | ||
| SBS | Chr5 | 23021902 | A-C | HET | ||
| SBS | Chr5 | 23640298 | C-A | HET | STOP_GAINED | LOC_Os05g40250 |
| SBS | Chr5 | 24006492 | G-A | HET | ||
| SBS | Chr5 | 4219100 | G-A | HET | ||
| SBS | Chr5 | 5038143 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g09070 |
| SBS | Chr5 | 7064636 | A-T | HET | ||
| SBS | Chr5 | 843459 | G-T | HET | ||
| SBS | Chr6 | 25451093 | G-C | HET | ||
| SBS | Chr6 | 8621751 | A-T | HET | ||
| SBS | Chr7 | 10068695 | C-G | HET | ||
| SBS | Chr7 | 14960388 | G-A | HET | ||
| SBS | Chr7 | 26562444 | A-G | HET | ||
| SBS | Chr7 | 7189420 | A-T | HET | ||
| SBS | Chr8 | 1058668 | T-C | Homo | ||
| SBS | Chr8 | 22110629 | C-T | HET | ||
| SBS | Chr8 | 22110630 | C-T | HET | ||
| SBS | Chr8 | 9221906 | A-G | Homo | ||
| SBS | Chr9 | 12967143 | G-A | Homo | ||
| SBS | Chr9 | 1656830 | G-C | HET | ||
| SBS | Chr9 | 2407599 | C-G | HET | ||
| SBS | Chr9 | 6566685 | G-A | HET | ||
| SBS | Chr9 | 9754910 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g15960 |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 749568 | 749570 | 2 | |
| Deletion | Chr10 | 2959284 | 2959288 | 4 | LOC_Os10g05860 |
| Deletion | Chr1 | 7821753 | 7821755 | 2 | |
| Deletion | Chr4 | 8484547 | 8484550 | 3 | |
| Deletion | Chr1 | 11106837 | 11106839 | 2 | LOC_Os01g19600 |
| Deletion | Chr2 | 11427579 | 11427581 | 2 | |
| Deletion | Chr2 | 12044542 | 12044545 | 3 | |
| Deletion | Chr2 | 12058839 | 12058866 | 27 | |
| Deletion | Chr2 | 12076132 | 12076145 | 13 | LOC_Os02g20470 |
| Deletion | Chr8 | 14144239 | 14144241 | 2 | LOC_Os08g23400 |
| Deletion | Chr4 | 14557154 | 14557164 | 10 | |
| Deletion | Chr5 | 15003510 | 15003514 | 4 | |
| Deletion | Chr4 | 15426316 | 15426317 | 1 | |
| Deletion | Chr2 | 16836846 | 16836855 | 9 | |
| Deletion | Chr1 | 17729787 | 17729791 | 4 | |
| Deletion | Chr11 | 18643597 | 18643599 | 2 | |
| Deletion | Chr12 | 20751813 | 20751826 | 13 | |
| Deletion | Chr6 | 21056718 | 21056720 | 2 | LOC_Os06g36020 |
| Deletion | Chr6 | 22276617 | 22276631 | 14 | |
| Deletion | Chr11 | 22367514 | 22367515 | 1 | LOC_Os11g37780 |
| Deletion | Chr8 | 23691429 | 23691439 | 10 | |
| Deletion | Chr5 | 23840423 | 23840424 | 1 | LOC_Os05g40650 |
| Deletion | Chr1 | 24736563 | 24736566 | 3 | |
| Deletion | Chr5 | 26513544 | 26513546 | 2 | LOC_Os05g45779 |
| Deletion | Chr12 | 26860186 | 26860187 | 1 | |
| Deletion | Chr5 | 27173528 | 27173530 | 2 | |
| Deletion | Chr11 | 27994283 | 27994284 | 1 | LOC_Os11g46230 |
| Deletion | Chr3 | 31738027 | 31738029 | 2 |
Insertions: 5
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 5161034 | 5413356 | 2 |
| Inversion | Chr7 | 12812272 | 13293673 | LOC_Os07g22720 |
| Inversion | Chr7 | 13306300 | 13719379 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 14290459 | Chr7 | 22952358 | 2 |
| Translocation | Chr4 | 20214654 | Chr1 | 11799235 |
