Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2283 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2283 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 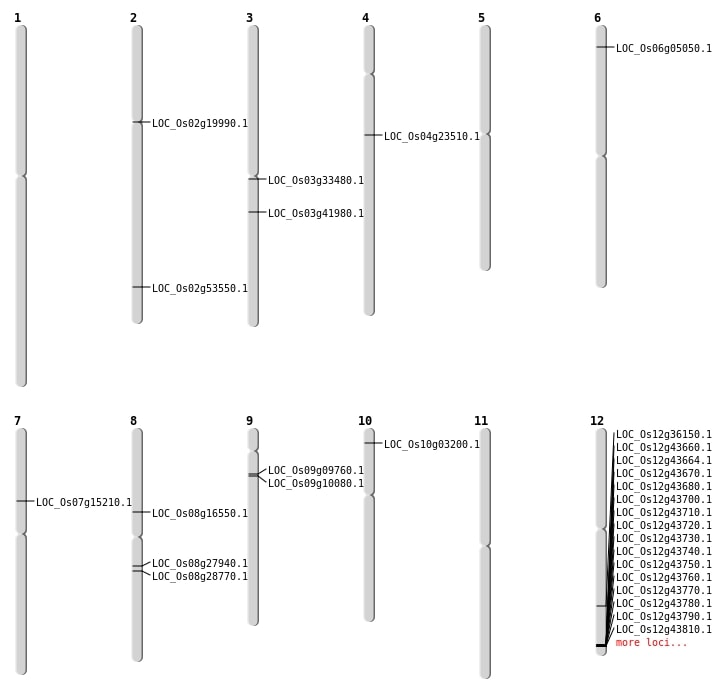
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 12326105 | T-C | HET | ||
| SBS | Chr10 | 1353092 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g03200 |
| SBS | Chr10 | 17150024 | C-T | HET | ||
| SBS | Chr10 | 9225829 | T-G | Homo | ||
| SBS | Chr11 | 11477660 | G-A | HET | ||
| SBS | Chr11 | 12742756 | T-C | HET | ||
| SBS | Chr11 | 17126274 | A-C | HET | ||
| SBS | Chr11 | 3904842 | A-G | HET | ||
| SBS | Chr12 | 10271759 | G-T | Homo | ||
| SBS | Chr12 | 22156139 | G-A | HET | STOP_GAINED | LOC_Os12g36150 |
| SBS | Chr12 | 23178514 | T-C | HET | ||
| SBS | Chr12 | 24162179 | C-G | HET | ||
| SBS | Chr12 | 7611130 | A-G | Homo | ||
| SBS | Chr2 | 32764606 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g53550 |
| SBS | Chr2 | 35883659 | G-C | HET | ||
| SBS | Chr3 | 19127897 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g33480 |
| SBS | Chr3 | 23324511 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g41980 |
| SBS | Chr3 | 8934073 | G-A | Homo | ||
| SBS | Chr4 | 10640225 | G-C | HET | ||
| SBS | Chr4 | 13439318 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g23510 |
| SBS | Chr4 | 27250874 | G-A | HET | ||
| SBS | Chr5 | 18278771 | A-T | HET | ||
| SBS | Chr5 | 19884892 | G-A | HET | ||
| SBS | Chr5 | 29772815 | C-T | Homo | ||
| SBS | Chr5 | 7210686 | G-A | HET | ||
| SBS | Chr5 | 9795742 | T-C | HET | ||
| SBS | Chr6 | 13019239 | G-C | Homo | ||
| SBS | Chr6 | 2232486 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g05050 |
| SBS | Chr7 | 11702965 | T-C | HET | ||
| SBS | Chr7 | 11702966 | A-C | HET | ||
| SBS | Chr7 | 18739681 | T-C | HET | ||
| SBS | Chr7 | 21447612 | C-T | HET | ||
| SBS | Chr7 | 6188866 | T-C | HET | ||
| SBS | Chr8 | 17195387 | G-A | HET | ||
| SBS | Chr8 | 17601925 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g28770 |
| SBS | Chr9 | 14756658 | G-A | Homo | ||
| SBS | Chr9 | 2655440 | C-T | HET | ||
| SBS | Chr9 | 5284467 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g09760 |
| SBS | Chr9 | 5495450 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g10080 |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1690613 | 1690639 | 26 | |
| Deletion | Chr7 | 2513601 | 2513610 | 9 | |
| Deletion | Chr4 | 3471697 | 3471705 | 8 | |
| Deletion | Chr3 | 7407866 | 7407873 | 7 | |
| Deletion | Chr10 | 7757140 | 7757156 | 16 | |
| Deletion | Chr5 | 7961516 | 7961518 | 2 | |
| Deletion | Chr3 | 12536117 | 12536118 | 1 | |
| Deletion | Chr11 | 13133298 | 13133300 | 2 | |
| Deletion | Chr8 | 17027936 | 17027939 | 3 | LOC_Os08g27940 |
| Deletion | Chr5 | 18036186 | 18036191 | 5 | |
| Deletion | Chr7 | 21335022 | 21335027 | 5 | |
| Deletion | Chr10 | 22288569 | 22288570 | 1 | |
| Deletion | Chr5 | 22803385 | 22803388 | 3 | |
| Deletion | Chr12 | 26284458 | 26284464 | 6 | |
| Deletion | Chr12 | 27124001 | 27166000 | 41999 | 8 |
| Deletion | Chr12 | 27169001 | 27259000 | 89999 | 17 |
| Deletion | Chr6 | 30832126 | 30832134 | 8 | |
| Deletion | Chr2 | 31885281 | 31885283 | 2 | |
| Deletion | Chr4 | 35074674 | 35074677 | 3 | |
| Deletion | Chr1 | 36188005 | 36188007 | 2 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr7 | 19065451 | 19065451 | 1 | |
| Insertion | Chr7 | 8766718 | 8766718 | 1 | LOC_Os07g15210 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 11748986 | 11760311 | 2 |
| Inversion | Chr2 | 11748991 | 11760308 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 10127990 | Chr4 | 8208152 | LOC_Os08g16550 |
