Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2304 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2304 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 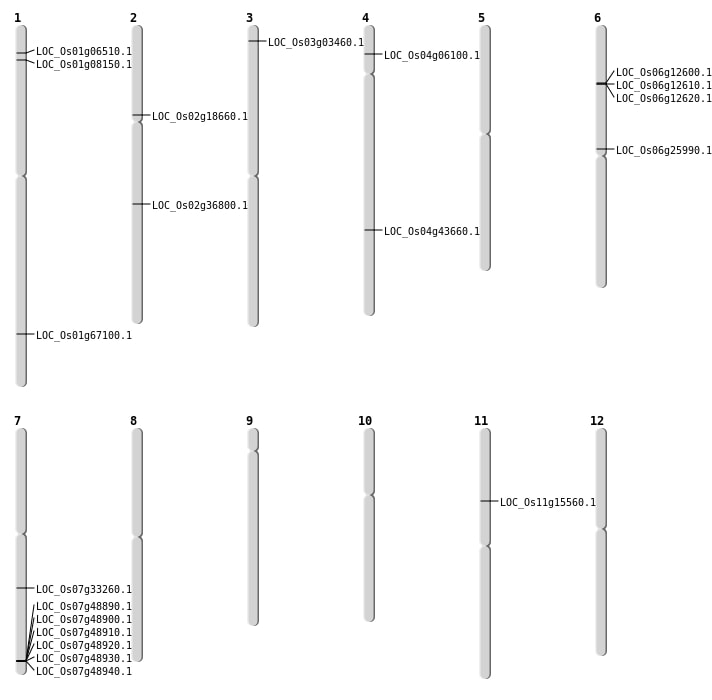
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15697978 | C-T | Homo | ||
| SBS | Chr1 | 22886957 | T-C | Homo | ||
| SBS | Chr1 | 26994345 | G-A | HET | ||
| SBS | Chr1 | 28962461 | C-T | Homo | ||
| SBS | Chr1 | 34054276 | G-A | HET | ||
| SBS | Chr1 | 34373943 | A-G | Homo | ||
| SBS | Chr1 | 37410634 | A-G | Homo | ||
| SBS | Chr1 | 38953723 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g67100 |
| SBS | Chr1 | 3941299 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g08150 |
| SBS | Chr1 | 3941300 | A-T | HET | ||
| SBS | Chr1 | 3964509 | A-G | HET | ||
| SBS | Chr10 | 11287194 | A-G | HET | ||
| SBS | Chr10 | 11287195 | G-A | HET | ||
| SBS | Chr10 | 19134489 | T-C | HET | ||
| SBS | Chr10 | 2054975 | C-T | HET | ||
| SBS | Chr10 | 4650378 | C-T | HET | ||
| SBS | Chr11 | 13768591 | T-C | Homo | ||
| SBS | Chr11 | 15376384 | C-G | HET | ||
| SBS | Chr11 | 20002590 | C-T | Homo | ||
| SBS | Chr11 | 8817025 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g15560 |
| SBS | Chr11 | 9589383 | C-T | HET | ||
| SBS | Chr12 | 13359822 | G-A | HET | ||
| SBS | Chr12 | 15572998 | C-T | HET | ||
| SBS | Chr12 | 8681893 | A-G | HET | ||
| SBS | Chr12 | 9002417 | G-A | HET | ||
| SBS | Chr2 | 10871050 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g18660 |
| SBS | Chr2 | 16124670 | C-T | HET | ||
| SBS | Chr2 | 22196895 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g36800 |
| SBS | Chr2 | 22973628 | T-C | HET | ||
| SBS | Chr3 | 28984926 | A-G | Homo | ||
| SBS | Chr3 | 8784677 | T-C | HET | ||
| SBS | Chr4 | 11234513 | T-A | HET | ||
| SBS | Chr4 | 13106460 | A-G | HET | ||
| SBS | Chr4 | 14547660 | G-A | Homo | ||
| SBS | Chr4 | 22477508 | C-A | Homo | ||
| SBS | Chr4 | 3168544 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g06100 |
| SBS | Chr5 | 16676579 | G-A | HET | ||
| SBS | Chr5 | 23837168 | G-A | HET | ||
| SBS | Chr5 | 720645 | C-T | HET | ||
| SBS | Chr5 | 8800546 | A-C | HET | ||
| SBS | Chr6 | 15394973 | T-C | HET | ||
| SBS | Chr6 | 16545264 | C-T | HET | ||
| SBS | Chr6 | 17990148 | T-C | HET | ||
| SBS | Chr6 | 26728015 | T-A | Homo | ||
| SBS | Chr6 | 26728019 | G-A | Homo | ||
| SBS | Chr7 | 13564181 | C-T | HET | ||
| SBS | Chr8 | 14487399 | C-T | HET | ||
| SBS | Chr8 | 20954182 | C-T | HET | ||
| SBS | Chr9 | 1732438 | C-T | HET | ||
| SBS | Chr9 | 9343285 | T-C | HET |
Deletions: 32
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 192672 | 192684 | 12 | |
| Deletion | Chr3 | 1496936 | 1496972 | 36 | LOC_Os03g03460 |
| Deletion | Chr5 | 2301679 | 2301683 | 4 | |
| Deletion | Chr1 | 2650176 | 2650342 | 166 | |
| Deletion | Chr1 | 3064131 | 3064133 | 2 | LOC_Os01g06510 |
| Deletion | Chr3 | 3551284 | 3551296 | 12 | |
| Deletion | Chr7 | 4055997 | 4055998 | 1 | |
| Deletion | Chr5 | 5117337 | 5117338 | 1 | |
| Deletion | Chr6 | 5308449 | 5308455 | 6 | |
| Deletion | Chr6 | 6837001 | 6873000 | 35999 | 2 |
| Deletion | Chr6 | 6874001 | 6885000 | 10999 | LOC_Os06g12620 |
| Deletion | Chr3 | 6921790 | 6921797 | 7 | |
| Deletion | Chr12 | 10813382 | 10813386 | 4 | |
| Deletion | Chr7 | 10954089 | 10954090 | 1 | |
| Deletion | Chr2 | 12310763 | 12310769 | 6 | |
| Deletion | Chr9 | 14104647 | 14104663 | 16 | |
| Deletion | Chr6 | 15219889 | 15219893 | 4 | LOC_Os06g25990 |
| Deletion | Chr5 | 15458053 | 15458073 | 20 | |
| Deletion | Chr9 | 16022951 | 16022954 | 3 | |
| Deletion | Chr12 | 17647108 | 17647109 | 1 | |
| Deletion | Chr7 | 19875693 | 19875698 | 5 | LOC_Os07g33260 |
| Deletion | Chr3 | 21666706 | 21666720 | 14 | |
| Deletion | Chr2 | 23397527 | 23397529 | 2 | |
| Deletion | Chr6 | 26547805 | 26547813 | 8 | |
| Deletion | Chr11 | 26706317 | 26706333 | 16 | |
| Deletion | Chr3 | 26960537 | 26960546 | 9 | |
| Deletion | Chr8 | 27625588 | 27625590 | 2 | |
| Deletion | Chr7 | 29255001 | 29283000 | 27999 | 6 |
| Deletion | Chr7 | 29403554 | 29403567 | 13 | |
| Deletion | Chr2 | 33762187 | 33762192 | 5 | |
| Deletion | Chr3 | 36091320 | 36091335 | 15 | |
| Deletion | Chr1 | 37487088 | 37487098 | 10 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 4548519 | 4548530 | 12 | |
| Insertion | Chr12 | 20273415 | 20273416 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 25833143 | 25904360 | LOC_Os04g43660 |
| Inversion | Chr4 | 25833158 | 25904370 | LOC_Os04g43660 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 1053180 | Chr4 | 31356447 | |
| Translocation | Chr5 | 1066713 | Chr4 | 31356461 | |
| Translocation | Chr10 | 9492675 | Chr4 | 13787584 |
