Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2310 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2310 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 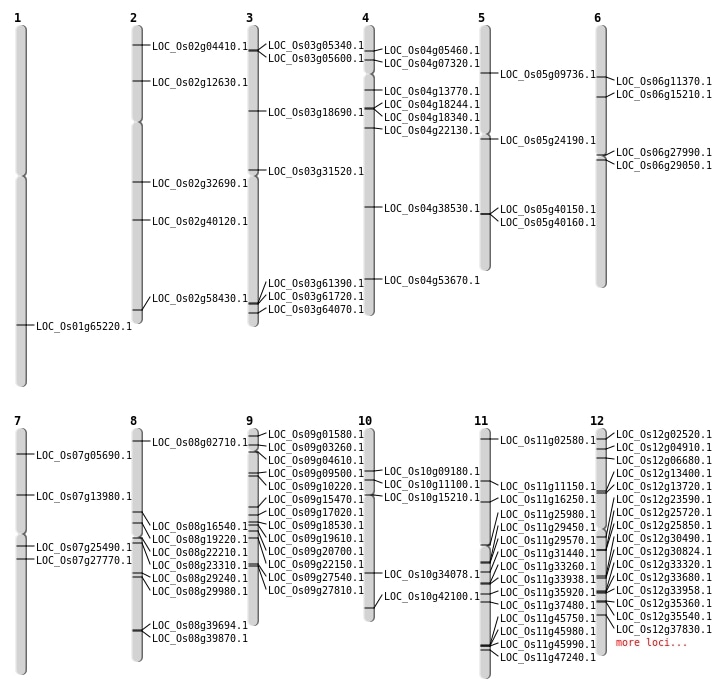
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14524636 | G-T | HET | ||
| SBS | Chr1 | 19928578 | A-T | HET | ||
| SBS | Chr1 | 29496420 | G-A | HET | ||
| SBS | Chr1 | 29637360 | G-A | HET | ||
| SBS | Chr1 | 36294000 | G-A | Homo | ||
| SBS | Chr1 | 8168293 | T-A | Homo | ||
| SBS | Chr10 | 15374369 | G-A | HET | ||
| SBS | Chr10 | 6791161 | T-C | HET | ||
| SBS | Chr11 | 19794340 | G-T | HET | ||
| SBS | Chr11 | 2097945 | A-T | HET | ||
| SBS | Chr12 | 16534900 | T-C | HET | ||
| SBS | Chr12 | 21260258 | G-A | HET | ||
| SBS | Chr12 | 53570 | C-T | HET | ||
| SBS | Chr2 | 14802054 | A-G | HET | ||
| SBS | Chr2 | 1840026 | C-T | HET | ||
| SBS | Chr2 | 19345531 | C-A | Homo | ||
| SBS | Chr2 | 19675958 | T-C | HET | ||
| SBS | Chr2 | 19917690 | C-T | Homo | ||
| SBS | Chr2 | 20112803 | G-T | Homo | ||
| SBS | Chr2 | 35734030 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g58430 |
| SBS | Chr2 | 6592312 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g12630 |
| SBS | Chr3 | 17957583 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g31520 |
| SBS | Chr3 | 18632222 | C-T | HET | ||
| SBS | Chr3 | 3115235 | G-C | HET | ||
| SBS | Chr3 | 36204771 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g64070 |
| SBS | Chr4 | 15502651 | C-A | HET | ||
| SBS | Chr5 | 12270422 | C-A | Homo | ||
| SBS | Chr5 | 14058897 | C-T | Homo | ||
| SBS | Chr5 | 18775362 | G-A | HET | ||
| SBS | Chr5 | 21511724 | G-T | HET | ||
| SBS | Chr6 | 12723313 | G-A | HET | ||
| SBS | Chr7 | 15905464 | C-G | HET | ||
| SBS | Chr7 | 16207228 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g27770 |
| SBS | Chr7 | 20356178 | C-A | HET | ||
| SBS | Chr7 | 27352032 | G-A | Homo | ||
| SBS | Chr9 | 14633705 | C-T | Homo | ||
| SBS | Chr9 | 18299845 | T-C | HET | ||
| SBS | Chr9 | 62718 | T-C | HET | ||
| SBS | Chr9 | 9008240 | C-A | HET |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 2082290 | 2082292 | 2 | |
| Deletion | Chr12 | 2098697 | 2098698 | 1 | |
| Deletion | Chr8 | 2899829 | 2899830 | 1 | |
| Deletion | Chr9 | 4735342 | 4735343 | 1 | |
| Deletion | Chr5 | 5523461 | 5523463 | 2 | LOC_Os05g09736 |
| Deletion | Chr1 | 6123390 | 6123391 | 1 | |
| Deletion | Chr3 | 7652399 | 7652405 | 6 | |
| Deletion | Chr12 | 8320007 | 8320011 | 4 | |
| Deletion | Chr4 | 10092927 | 10092932 | 5 | LOC_Os04g18244 |
| Deletion | Chr8 | 14073558 | 14073560 | 2 | LOC_Os08g23310 |
| Deletion | Chr7 | 16561356 | 16561375 | 19 | |
| Deletion | Chr11 | 17093400 | 17093407 | 7 | LOC_Os11g29450 |
| Deletion | Chr2 | 19871679 | 19871687 | 8 | |
| Deletion | Chr5 | 23864692 | 23864697 | 5 | |
| Deletion | Chr11 | 25123425 | 25123449 | 24 | |
| Deletion | Chr1 | 26345883 | 26345886 | 3 | |
| Deletion | Chr2 | 27902948 | 27902949 | 1 | |
| Deletion | Chr1 | 31616852 | 31616879 | 27 | |
| Deletion | Chr3 | 34841609 | 34841610 | 1 | LOC_Os03g61390 |
Insertions: 6
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 9480227 | 9501939 | |
| Inversion | Chr12 | 9480250 | 9501952 | |
| Inversion | Chr4 | 29509156 | 30289943 | |
| Inversion | Chr4 | 29509166 | 30289953 | |
| Inversion | Chr3 | 34986408 | 34986537 | LOC_Os03g61720 |
Translocations: 135
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 858416 | Chr11 | 815528 | 2 |
| Translocation | Chr12 | 2109733 | Chr9 | 9469239 | 2 |
| Translocation | Chr12 | 2147144 | Chr11 | 5073923 | |
| Translocation | Chr11 | 2153913 | Chr9 | 8955408 | |
| Translocation | Chr11 | 2163974 | Chr1 | 37409969 | |
| Translocation | Chr4 | 3220802 | Chr2 | 19314573 | |
| Translocation | Chr12 | 3253819 | Chr11 | 21096707 | 2 |
| Translocation | Chr12 | 3699304 | Chr4 | 9565765 | |
| Translocation | Chr7 | 3981617 | Chr3 | 2616130 | 2 |
| Translocation | Chr10 | 4089543 | Chr6 | 14634242 | |
| Translocation | Chr10 | 4210026 | Chr7 | 5465962 | |
| Translocation | Chr12 | 4416697 | Chr8 | 18415250 | 2 |
| Translocation | Chr9 | 5120596 | Chr4 | 3943172 | LOC_Os09g09500 |
| Translocation | Chr11 | 5123058 | Chr2 | 1945054 | 2 |
| Translocation | Chr10 | 5623931 | Chr6 | 8458367 | |
| Translocation | Chr12 | 5779287 | Chr8 | 19753812 | |
| Translocation | Chr8 | 5977028 | Chr3 | 2803169 | 2 |
| Translocation | Chr6 | 5978743 | Chr4 | 3687509 | LOC_Os06g11370 |
| Translocation | Chr12 | 6248354 | Chr8 | 4190745 | |
| Translocation | Chr9 | 6282553 | Chr4 | 5870526 | |
| Translocation | Chr12 | 6808366 | Chr11 | 13095623 | |
| Translocation | Chr10 | 6900294 | Chr5 | 2479642 | |
| Translocation | Chr9 | 7389118 | Chr3 | 6343122 | |
| Translocation | Chr8 | 7455821 | Chr2 | 16612646 | |
| Translocation | Chr12 | 7498166 | Chr7 | 13571132 | LOC_Os12g13400 |
| Translocation | Chr12 | 7742402 | Chr2 | 19414035 | 2 |
| Translocation | Chr5 | 7847652 | Chr3 | 23251985 | |
| Translocation | Chr10 | 8053110 | Chr7 | 15100151 | LOC_Os10g15210 |
| Translocation | Chr3 | 8825038 | Chr2 | 19352215 | |
| Translocation | Chr11 | 9355078 | Chr8 | 9822301 | |
| Translocation | Chr12 | 9754251 | Chr10 | 8039778 | |
| Translocation | Chr12 | 10087162 | Chr8 | 21909928 | |
| Translocation | Chr6 | 10144060 | Chr3 | 22322171 | |
| Translocation | Chr7 | 10511348 | Chr6 | 11334670 | |
| Translocation | Chr11 | 10756223 | Chr4 | 10128571 | 2 |
| Translocation | Chr12 | 11561161 | Chr2 | 29427397 | |
| Translocation | Chr9 | 11597072 | Chr6 | 10212962 | |
| Translocation | Chr4 | 11694426 | Chr3 | 12620075 | |
| Translocation | Chr9 | 11727109 | Chr4 | 17810694 | LOC_Os09g19610 |
| Translocation | Chr9 | 12466633 | Chr8 | 8352631 | LOC_Os09g20700 |
| Translocation | Chr12 | 12473328 | Chr9 | 5559711 | |
| Translocation | Chr11 | 13169663 | Chr9 | 2457129 | 2 |
| Translocation | Chr11 | 13306791 | Chr4 | 31987972 | 2 |
| Translocation | Chr12 | 13371990 | Chr10 | 16479593 | LOC_Os12g23590 |
| Translocation | Chr12 | 13373658 | Chr4 | 12545924 | 2 |
| Translocation | Chr8 | 13399778 | Chr7 | 7949080 | LOC_Os08g22210 |
| Translocation | Chr9 | 13409018 | Chr6 | 18842977 | LOC_Os09g22150 |
| Translocation | Chr11 | 13528941 | Chr10 | 6141895 | 2 |
| Translocation | Chr12 | 13614721 | Chr7 | 6553016 | |
| Translocation | Chr8 | 13663382 | Chr5 | 22889167 | |
| Translocation | Chr12 | 14086274 | Chr8 | 10124017 | 2 |
| Translocation | Chr12 | 14087109 | Chr11 | 8944378 | 2 |
| Translocation | Chr10 | 14392105 | Chr6 | 13904099 | |
| Translocation | Chr8 | 14737228 | Chr7 | 5214938 | |
| Translocation | Chr9 | 14795702 | Chr7 | 5461185 | |
| Translocation | Chr12 | 14916315 | Chr9 | 5569626 | 2 |
| Translocation | Chr9 | 14951257 | Chr1 | 37409863 | |
| Translocation | Chr12 | 14995788 | Chr11 | 6169092 | 2 |
| Translocation | Chr10 | 15557603 | Chr4 | 15030259 | |
| Translocation | Chr7 | 15625895 | Chr1 | 3307639 | |
| Translocation | Chr8 | 15901291 | Chr4 | 7677717 | 2 |
| Translocation | Chr12 | 16085337 | Chr9 | 11362256 | 2 |
| Translocation | Chr12 | 16204352 | Chr4 | 3633936 | |
| Translocation | Chr8 | 16263154 | Chr6 | 15882910 | 2 |
| Translocation | Chr10 | 16394812 | Chr9 | 19091217 | |
| Translocation | Chr6 | 16552093 | Chr4 | 2748318 | 2 |
| Translocation | Chr11 | 16694121 | Chr7 | 9894221 | |
| Translocation | Chr9 | 16737773 | Chr7 | 1125471 | LOC_Os09g27540 |
| Translocation | Chr8 | 16783131 | Chr5 | 13970147 | 2 |
| Translocation | Chr10 | 16821028 | Chr1 | 41587176 | |
| Translocation | Chr9 | 16923207 | Chr4 | 13705017 | LOC_Os09g27810 |
| Translocation | Chr12 | 16959285 | Chr8 | 6751661 | |
| Translocation | Chr11 | 17158377 | Chr10 | 22891075 | LOC_Os11g29570 |
| Translocation | Chr11 | 17701376 | Chr8 | 24046280 | |
| Translocation | Chr7 | 18180329 | Chr4 | 3889343 | 2 |
| Translocation | Chr10 | 18189458 | Chr9 | 1568515 | 2 |
| Translocation | Chr12 | 18301853 | Chr9 | 21567476 | LOC_Os12g30490 |
| Translocation | Chr9 | 18370188 | Chr8 | 2960408 | |
| Translocation | Chr11 | 18371992 | Chr10 | 879842 | LOC_Os11g31440 |
| Translocation | Chr11 | 18508356 | Chr1 | 40950870 | |
| Translocation | Chr12 | 18521475 | Chr9 | 1654382 | LOC_Os12g30824 |
| Translocation | Chr8 | 18745849 | Chr3 | 18929781 | |
| Translocation | Chr11 | 19660583 | Chr7 | 2719536 | 2 |
| Translocation | Chr11 | 19843562 | Chr1 | 19348796 | LOC_Os11g33938 |
| Translocation | Chr12 | 20096547 | Chr6 | 23394909 | |
| Translocation | Chr12 | 20149124 | Chr10 | 4958961 | 2 |
| Translocation | Chr12 | 20153354 | Chr11 | 14826899 | 2 |
| Translocation | Chr12 | 20220322 | Chr4 | 271747 | |
| Translocation | Chr6 | 20282799 | Chr5 | 6098939 | |
| Translocation | Chr12 | 20331815 | Chr8 | 17926417 | 2 |
| Translocation | Chr4 | 20346294 | Chr3 | 5807536 | |
| Translocation | Chr12 | 20507508 | Chr1 | 33160323 | LOC_Os12g33958 |
| Translocation | Chr12 | 20507518 | Chr6 | 4239147 | LOC_Os12g33958 |
| Translocation | Chr12 | 20542616 | Chr1 | 37847717 | 2 |
| Translocation | Chr12 | 21142573 | Chr9 | 5132484 | |
| Translocation | Chr7 | 21174152 | Chr4 | 8324312 | |
| Translocation | Chr6 | 21200642 | Chr4 | 8543233 | |
| Translocation | Chr12 | 21494618 | Chr11 | 17511560 | LOC_Os12g35360 |
| Translocation | Chr11 | 21589579 | Chr7 | 7985056 | 2 |
| Translocation | Chr12 | 21609233 | Chr5 | 7138587 | LOC_Os12g35540 |
| Translocation | Chr12 | 21931774 | Chr2 | 2418114 | |
| Translocation | Chr11 | 22131076 | Chr6 | 15401537 | LOC_Os11g37480 |
| Translocation | Chr11 | 22180241 | Chr5 | 16082507 | |
| Translocation | Chr11 | 22362487 | Chr10 | 15002739 | |
| Translocation | Chr8 | 22364949 | Chr2 | 19181603 | |
| Translocation | Chr10 | 22644007 | Chr8 | 19750044 | LOC_Os10g42100 |
| Translocation | Chr4 | 22896135 | Chr2 | 13397069 | LOC_Os04g38530 |
| Translocation | Chr11 | 23032332 | Chr3 | 16389005 | |
| Translocation | Chr12 | 23226088 | Chr9 | 10415104 | 2 |
| Translocation | Chr12 | 23236491 | Chr8 | 4010297 | LOC_Os12g37830 |
| Translocation | Chr12 | 23542986 | Chr6 | 21994034 | |
| Translocation | Chr6 | 23608285 | Chr2 | 20852147 | |
| Translocation | Chr12 | 23803684 | Chr3 | 22077522 | |
| Translocation | Chr8 | 24365118 | Chr2 | 24289050 | 2 |
| Translocation | Chr12 | 24575513 | Chr3 | 18534145 | |
| Translocation | Chr11 | 24830135 | Chr10 | 20543868 | |
| Translocation | Chr8 | 25127054 | Chr6 | 8619512 | 2 |
| Translocation | Chr12 | 25194111 | Chr8 | 11496200 | 2 |
| Translocation | Chr4 | 25205774 | Chr3 | 16530742 | |
| Translocation | Chr8 | 25261237 | Chr7 | 14604277 | 2 |
| Translocation | Chr7 | 25285592 | Chr2 | 13402391 | |
| Translocation | Chr5 | 25539359 | Chr1 | 40623925 | |
| Translocation | Chr12 | 25668908 | Chr2 | 9937755 | |
| Translocation | Chr12 | 26482352 | Chr8 | 1132862 | 2 |
| Translocation | Chr12 | 27005780 | Chr2 | 10461207 | |
| Translocation | Chr11 | 27569194 | Chr5 | 16490292 | |
| Translocation | Chr5 | 27683279 | Chr3 | 10462479 | 2 |
| Translocation | Chr11 | 27688464 | Chr5 | 23584240 | 2 |
| Translocation | Chr7 | 27769966 | Chr1 | 29676763 | |
| Translocation | Chr11 | 27820948 | Chr5 | 23587093 | 2 |
| Translocation | Chr11 | 27827436 | Chr5 | 15902468 | |
| Translocation | Chr11 | 27831690 | Chr7 | 3937360 | LOC_Os11g45990 |
| Translocation | Chr11 | 28408444 | Chr9 | 408710 | 2 |
| Translocation | Chr3 | 28717934 | Chr2 | 6171094 | |
| Translocation | Chr11 | 28753002 | Chr1 | 28104454 |
