Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2333 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2333 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 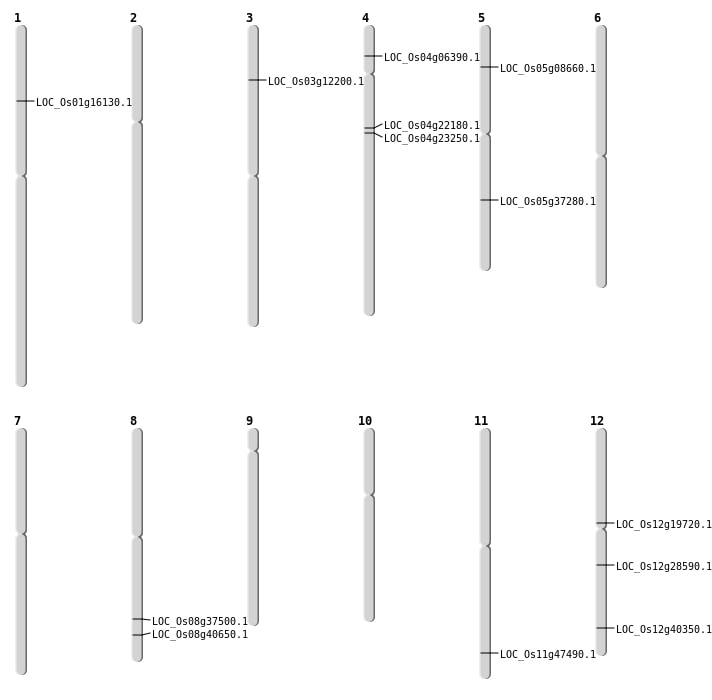
|
| Variant Information |
|---|
Single base substitutions: 56
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12693421 | G-A | HET | ||
| SBS | Chr1 | 18263342 | T-A | HET | ||
| SBS | Chr1 | 20510468 | T-A | HET | ||
| SBS | Chr1 | 21434586 | A-G | HET | ||
| SBS | Chr1 | 2248648 | T-C | HET | ||
| SBS | Chr1 | 27711114 | C-T | Homo | ||
| SBS | Chr1 | 37140262 | G-A | HET | ||
| SBS | Chr1 | 6865103 | G-A | HET | ||
| SBS | Chr10 | 23100089 | G-A | HET | ||
| SBS | Chr10 | 2484291 | G-A | Homo | ||
| SBS | Chr11 | 13031883 | A-C | HET | ||
| SBS | Chr11 | 15494552 | G-A | HET | ||
| SBS | Chr11 | 15494553 | T-A | HET | ||
| SBS | Chr11 | 16644120 | G-T | HET | ||
| SBS | Chr11 | 21717696 | A-T | HET | ||
| SBS | Chr11 | 2680485 | T-A | HET | ||
| SBS | Chr12 | 10980154 | C-T | HET | ||
| SBS | Chr12 | 13410332 | G-C | HET | ||
| SBS | Chr12 | 16909518 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g28590 |
| SBS | Chr12 | 23978414 | T-C | HET | ||
| SBS | Chr12 | 24984703 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g40350 |
| SBS | Chr12 | 2639277 | A-C | HET | ||
| SBS | Chr2 | 2185883 | G-A | HET | ||
| SBS | Chr2 | 35137403 | G-A | HET | ||
| SBS | Chr2 | 35141013 | G-T | HET | ||
| SBS | Chr3 | 1161476 | C-T | HET | ||
| SBS | Chr3 | 21260426 | C-T | HET | ||
| SBS | Chr3 | 2982815 | T-C | HET | ||
| SBS | Chr3 | 31582080 | C-T | HET | ||
| SBS | Chr4 | 12561182 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g22180 |
| SBS | Chr4 | 13188442 | C-A | HET | ||
| SBS | Chr4 | 13188443 | T-A | HET | ||
| SBS | Chr4 | 13266929 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g23250 |
| SBS | Chr4 | 22251145 | G-A | HET | ||
| SBS | Chr4 | 3384925 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g06390 |
| SBS | Chr5 | 21813244 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g37280 |
| SBS | Chr5 | 2262967 | C-A | Homo | ||
| SBS | Chr5 | 4664870 | G-C | HET | ||
| SBS | Chr6 | 11554860 | C-T | HET | ||
| SBS | Chr6 | 12251124 | G-A | HET | ||
| SBS | Chr6 | 26818053 | T-A | HET | ||
| SBS | Chr6 | 3734305 | C-T | HET | ||
| SBS | Chr6 | 8358187 | A-T | HET | ||
| SBS | Chr7 | 13507882 | G-A | HET | ||
| SBS | Chr7 | 21138823 | C-A | HET | ||
| SBS | Chr7 | 6951679 | A-T | HET | ||
| SBS | Chr8 | 12049868 | C-T | HET | ||
| SBS | Chr8 | 1639126 | G-T | HET | ||
| SBS | Chr8 | 18725648 | A-T | HET | ||
| SBS | Chr8 | 1956372 | G-A | HET | ||
| SBS | Chr8 | 21255507 | T-G | HET | ||
| SBS | Chr8 | 23757792 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g37500 |
| SBS | Chr8 | 3977511 | T-C | HET | ||
| SBS | Chr9 | 16039480 | C-A | HET | ||
| SBS | Chr9 | 21201415 | G-A | HET | ||
| SBS | Chr9 | 375587 | C-A | HET |
Deletions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 6399115 | 6399116 | 1 | LOC_Os03g12200 |
| Deletion | Chr12 | 11487340 | 11487341 | 1 | LOC_Os12g19720 |
| Deletion | Chr2 | 21596902 | 21596912 | 10 | |
| Deletion | Chr10 | 22493405 | 22493406 | 1 | |
| Deletion | Chr10 | 23029945 | 23029948 | 3 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 1476412 | 1476413 | 2 | |
| Insertion | Chr5 | 4740328 | 4740357 | 30 | LOC_Os05g08660 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 33253574 | 33858517 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 10914692 | Chr5 | 8834671 | |
| Translocation | Chr8 | 12316674 | Chr1 | 9100397 | 2 |
| Translocation | Chr8 | 15934975 | Chr7 | 6243005 | |
| Translocation | Chr10 | 16394933 | Chr4 | 27332032 | |
| Translocation | Chr12 | 27530417 | Chr8 | 7729489 | |
| Translocation | Chr11 | 28718626 | Chr8 | 25742856 | 2 |
