Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2373 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2373 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 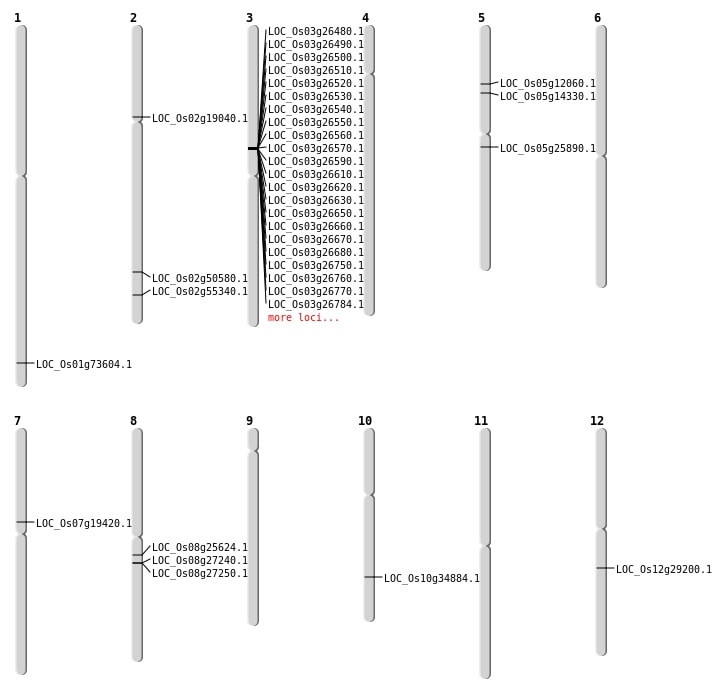
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10520522 | C-T | HET | ||
| SBS | Chr1 | 10520523 | C-T | HET | ||
| SBS | Chr1 | 19359100 | G-A | Homo | ||
| SBS | Chr1 | 32718221 | C-T | Homo | ||
| SBS | Chr1 | 34784564 | C-A | HET | ||
| SBS | Chr1 | 42639090 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g73604 |
| SBS | Chr10 | 11509126 | C-T | HET | ||
| SBS | Chr10 | 4896881 | G-A | HET | ||
| SBS | Chr11 | 19309368 | G-A | HET | ||
| SBS | Chr12 | 197359 | A-G | Homo | ||
| SBS | Chr12 | 8171949 | A-G | Homo | ||
| SBS | Chr2 | 11119345 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g19040 |
| SBS | Chr2 | 20212545 | G-A | HET | ||
| SBS | Chr2 | 30891321 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g50580 |
| SBS | Chr2 | 33893425 | A-T | Homo | SPLICE_SITE_DONOR | LOC_Os02g55340 |
| SBS | Chr3 | 14465505 | G-A | HET | ||
| SBS | Chr3 | 23731305 | T-A | Homo | ||
| SBS | Chr4 | 18908713 | G-T | HET | ||
| SBS | Chr4 | 2364352 | G-T | HET | ||
| SBS | Chr4 | 25214672 | C-A | HET | ||
| SBS | Chr4 | 25214673 | C-A | HET | ||
| SBS | Chr4 | 35495575 | G-A | HET | ||
| SBS | Chr4 | 5402879 | G-A | HET | ||
| SBS | Chr5 | 20977566 | T-C | HET | ||
| SBS | Chr5 | 3308318 | T-C | HET | ||
| SBS | Chr5 | 6897718 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g12060 |
| SBS | Chr5 | 8059539 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g14330 |
| SBS | Chr6 | 27631474 | A-C | Homo | ||
| SBS | Chr6 | 27730769 | T-A | Homo | ||
| SBS | Chr6 | 3028477 | C-A | HET | ||
| SBS | Chr7 | 11489776 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g19420 |
| SBS | Chr7 | 2458484 | T-G | Homo | ||
| SBS | Chr7 | 8569402 | T-A | HET | ||
| SBS | Chr8 | 10865340 | C-A | HET | ||
| SBS | Chr8 | 15592727 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g25624 |
| SBS | Chr8 | 16646364 | G-A | HET | ||
| SBS | Chr8 | 16646365 | A-G | HET | ||
| SBS | Chr8 | 19419834 | G-A | HET | ||
| SBS | Chr8 | 22545448 | A-G | HET | ||
| SBS | Chr8 | 22701182 | G-A | HET | ||
| SBS | Chr9 | 10227219 | A-G | HET | ||
| SBS | Chr9 | 13827511 | T-C | HET | ||
| SBS | Chr9 | 17220086 | C-T | HET | ||
| SBS | Chr9 | 21761719 | T-C | HET | ||
| SBS | Chr9 | 6174853 | C-T | HET |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 666147 | 666177 | 30 | |
| Deletion | Chr5 | 1441119 | 1441123 | 4 | |
| Deletion | Chr1 | 4634785 | 4634790 | 5 | |
| Deletion | Chr1 | 5451380 | 5451381 | 1 | |
| Deletion | Chr8 | 5840197 | 5840199 | 2 | |
| Deletion | Chr11 | 7636587 | 7636602 | 15 | |
| Deletion | Chr1 | 7815419 | 7815420 | 1 | |
| Deletion | Chr3 | 13028563 | 13028566 | 3 | |
| Deletion | Chr5 | 15065861 | 15065872 | 11 | LOC_Os05g25890 |
| Deletion | Chr3 | 15124001 | 15237000 | 112999 | 18 |
| Deletion | Chr3 | 15259001 | 15312000 | 52999 | 9 |
| Deletion | Chr8 | 16624001 | 16643000 | 18999 | 2 |
| Deletion | Chr7 | 16726031 | 16726032 | 1 | |
| Deletion | Chr12 | 17292404 | 17292406 | 2 | LOC_Os12g29200 |
| Deletion | Chr10 | 18248111 | 18248118 | 7 | |
| Deletion | Chr10 | 18616806 | 18616807 | 1 | LOC_Os10g34884 |
| Deletion | Chr2 | 20310935 | 20310936 | 1 | |
| Deletion | Chr5 | 20778935 | 20778936 | 1 | |
| Deletion | Chr5 | 23287550 | 23287554 | 4 | |
| Deletion | Chr4 | 24124537 | 24124539 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 20536395 | 20536396 | 2 | |
| Insertion | Chr12 | 17639261 | 17639262 | 2 | |
| Insertion | Chr9 | 13061771 | 13061772 | 2 |
No Inversion
No Translocation
