Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2378 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2378 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 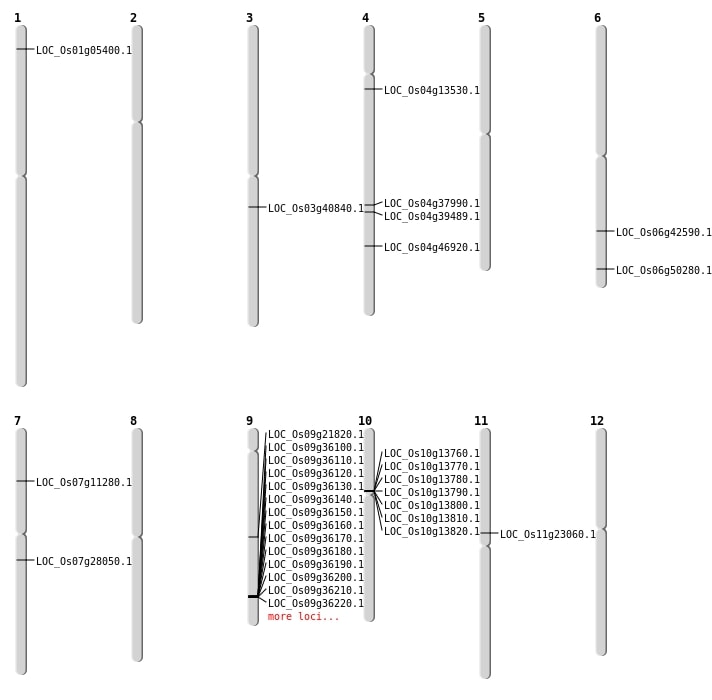
|
| Variant Information |
|---|
Single base substitutions: 63
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1215433 | T-G | HET | ||
| SBS | Chr1 | 1215482 | T-C | HET | ||
| SBS | Chr1 | 13490788 | C-T | HET | ||
| SBS | Chr1 | 20623911 | C-A | HET | ||
| SBS | Chr10 | 2423889 | T-A | Homo | ||
| SBS | Chr10 | 2423890 | T-A | Homo | ||
| SBS | Chr10 | 246348 | T-G | HET | ||
| SBS | Chr10 | 7692097 | T-C | HET | ||
| SBS | Chr10 | 8913414 | C-A | HET | ||
| SBS | Chr11 | 12293212 | T-G | HET | ||
| SBS | Chr11 | 12931203 | C-T | HET | ||
| SBS | Chr11 | 13270680 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g23060 |
| SBS | Chr11 | 16083527 | G-A | HET | ||
| SBS | Chr11 | 20555135 | A-T | HET | ||
| SBS | Chr12 | 13946969 | C-T | HET | ||
| SBS | Chr12 | 19331957 | C-T | HET | ||
| SBS | Chr12 | 3631186 | G-A | Homo | ||
| SBS | Chr12 | 5788286 | T-A | HET | ||
| SBS | Chr12 | 9095080 | C-T | HET | ||
| SBS | Chr12 | 9585570 | T-C | HET | ||
| SBS | Chr2 | 1930294 | C-T | HET | ||
| SBS | Chr2 | 2174147 | G-T | HET | ||
| SBS | Chr2 | 31262796 | G-T | Homo | ||
| SBS | Chr2 | 3959736 | T-A | HET | ||
| SBS | Chr2 | 4648013 | T-A | HET | ||
| SBS | Chr2 | 8103032 | T-G | HET | ||
| SBS | Chr3 | 17176129 | C-T | HET | ||
| SBS | Chr3 | 19718748 | C-T | HET | ||
| SBS | Chr3 | 22713109 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g40840 |
| SBS | Chr3 | 25024031 | G-A | HET | ||
| SBS | Chr3 | 31275346 | A-G | HET | ||
| SBS | Chr4 | 11480130 | A-T | Homo | ||
| SBS | Chr4 | 13480539 | C-A | HET | ||
| SBS | Chr4 | 13702502 | A-T | Homo | ||
| SBS | Chr4 | 14258106 | T-A | Homo | ||
| SBS | Chr4 | 23394592 | A-T | HET | ||
| SBS | Chr4 | 24511309 | C-T | HET | ||
| SBS | Chr4 | 31369778 | C-T | HET | ||
| SBS | Chr4 | 33318010 | A-T | HET | ||
| SBS | Chr4 | 3433104 | G-A | HET | ||
| SBS | Chr4 | 9871927 | C-T | HET | ||
| SBS | Chr6 | 21560935 | A-G | HET | ||
| SBS | Chr6 | 25614011 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g42590 |
| SBS | Chr6 | 2627441 | C-A | HET | ||
| SBS | Chr6 | 27967040 | G-A | Homo | ||
| SBS | Chr6 | 27967041 | C-A | Homo | ||
| SBS | Chr6 | 30435344 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g50280 |
| SBS | Chr7 | 13297798 | G-C | HET | ||
| SBS | Chr7 | 14257916 | A-G | HET | ||
| SBS | Chr7 | 14322511 | T-A | HET | ||
| SBS | Chr7 | 16359730 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g28050 |
| SBS | Chr7 | 4226226 | A-G | Homo | ||
| SBS | Chr7 | 6228215 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g11280 |
| SBS | Chr7 | 7828771 | C-T | HET | ||
| SBS | Chr7 | 8752538 | G-A | HET | ||
| SBS | Chr7 | 9918570 | G-A | HET | ||
| SBS | Chr8 | 23139691 | C-T | HET | ||
| SBS | Chr8 | 4879497 | A-G | HET | ||
| SBS | Chr8 | 526786 | G-A | HET | ||
| SBS | Chr8 | 8158844 | A-T | HET | ||
| SBS | Chr9 | 13224964 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g21820 |
| SBS | Chr9 | 13874931 | T-A | HET | ||
| SBS | Chr9 | 18290620 | T-C | HET |
Deletions: 19
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 5481470 | 5481475 | 6 | |
| Insertion | Chr6 | 27114887 | 27114888 | 2 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 22618786 | 23512746 | 2 |
| Inversion | Chr4 | 22618791 | 23512751 | 2 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 7547480 | Chr1 | 2551141 | 2 |
| Translocation | Chr6 | 10194145 | Chr2 | 30977614 | |
| Translocation | Chr8 | 11738402 | Chr4 | 27822177 | 2 |
| Translocation | Chr8 | 11738770 | Chr4 | 27822180 | 2 |
