Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2386 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2386 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 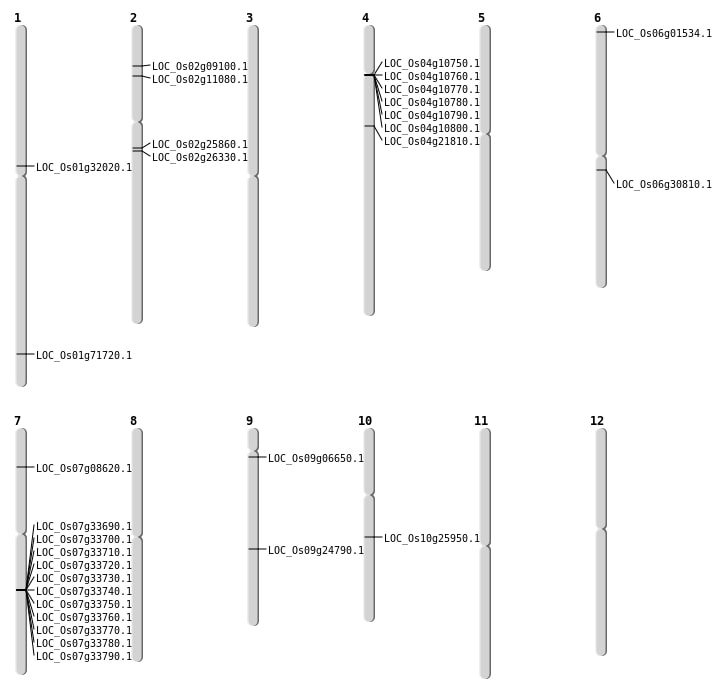
|
| Variant Information |
|---|
Single base substitutions: 71
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13844400 | G-T | HET | ||
| SBS | Chr1 | 16627004 | A-G | Homo | ||
| SBS | Chr1 | 17532252 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g32020 |
| SBS | Chr1 | 21400807 | G-A | HET | ||
| SBS | Chr1 | 30559070 | T-C | HET | ||
| SBS | Chr1 | 3359646 | G-A | HET | ||
| SBS | Chr1 | 35109184 | A-T | HET | ||
| SBS | Chr1 | 35172488 | A-G | HET | ||
| SBS | Chr1 | 35721015 | C-T | HET | ||
| SBS | Chr1 | 37672115 | C-A | HET | ||
| SBS | Chr10 | 13450091 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g25950 |
| SBS | Chr10 | 8637171 | G-A | HET | ||
| SBS | Chr11 | 17333300 | C-G | HET | ||
| SBS | Chr11 | 833034 | G-T | HET | ||
| SBS | Chr12 | 17458860 | A-G | HET | ||
| SBS | Chr12 | 1754825 | G-A | HET | ||
| SBS | Chr12 | 24302599 | G-A | HET | ||
| SBS | Chr12 | 24833183 | C-A | HET | ||
| SBS | Chr12 | 498911 | G-T | HET | ||
| SBS | Chr12 | 7445111 | A-T | HET | ||
| SBS | Chr12 | 8832115 | C-T | HET | ||
| SBS | Chr2 | 34554251 | A-G | HET | ||
| SBS | Chr2 | 4668547 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g09100 |
| SBS | Chr2 | 5936896 | G-T | HET | STOP_GAINED | LOC_Os02g11080 |
| SBS | Chr2 | 5936901 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g11080 |
| SBS | Chr3 | 1189750 | G-A | HET | ||
| SBS | Chr3 | 17648221 | G-A | HET | ||
| SBS | Chr3 | 22405400 | G-A | HET | ||
| SBS | Chr3 | 27182532 | G-A | HET | ||
| SBS | Chr3 | 29822671 | T-C | HET | ||
| SBS | Chr3 | 3090057 | T-C | HET | ||
| SBS | Chr3 | 32113140 | C-A | HET | ||
| SBS | Chr3 | 4574153 | A-G | HET | ||
| SBS | Chr3 | 6464149 | T-C | HET | ||
| SBS | Chr3 | 9890171 | C-T | HET | ||
| SBS | Chr4 | 10465734 | G-A | HET | ||
| SBS | Chr4 | 12359133 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g21810 |
| SBS | Chr4 | 13195371 | G-A | HET | ||
| SBS | Chr4 | 18471633 | C-T | HET | ||
| SBS | Chr4 | 26039374 | G-C | HET | ||
| SBS | Chr4 | 320983 | A-T | Homo | ||
| SBS | Chr4 | 4710278 | A-G | HET | ||
| SBS | Chr4 | 6321603 | G-T | HET | ||
| SBS | Chr5 | 11869280 | C-A | Homo | ||
| SBS | Chr5 | 11869283 | A-T | Homo | ||
| SBS | Chr5 | 25827611 | C-T | HET | ||
| SBS | Chr5 | 4410459 | C-T | HET | ||
| SBS | Chr6 | 17869553 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g30810 |
| SBS | Chr6 | 479116 | A-G | HET | ||
| SBS | Chr7 | 16160806 | G-A | HET | ||
| SBS | Chr7 | 16769309 | C-T | Homo | ||
| SBS | Chr7 | 16769310 | A-T | Homo | ||
| SBS | Chr7 | 16808172 | C-T | HET | ||
| SBS | Chr7 | 17006162 | G-A | HET | ||
| SBS | Chr7 | 18839092 | T-C | HET | ||
| SBS | Chr7 | 20631875 | G-C | HET | ||
| SBS | Chr7 | 25331098 | C-T | HET | ||
| SBS | Chr7 | 26115917 | G-A | HET | ||
| SBS | Chr7 | 27444984 | G-A | Homo | ||
| SBS | Chr7 | 4443183 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g08620 |
| SBS | Chr8 | 13123773 | G-C | HET | ||
| SBS | Chr8 | 18124384 | T-C | HET | ||
| SBS | Chr8 | 23361473 | T-C | HET | ||
| SBS | Chr8 | 25516915 | T-C | HET | ||
| SBS | Chr8 | 6379182 | G-C | HET | ||
| SBS | Chr8 | 9779166 | T-C | HET | ||
| SBS | Chr9 | 10319898 | C-A | HET | ||
| SBS | Chr9 | 14764626 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g24790 |
| SBS | Chr9 | 17927268 | A-G | HET | ||
| SBS | Chr9 | 3160536 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g06650 |
| SBS | Chr9 | 4984183 | G-A | HET |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 325162 | 325163 | 1 | LOC_Os06g01534 |
| Deletion | Chr4 | 1910620 | 1910621 | 1 | |
| Deletion | Chr5 | 3802872 | 3802901 | 29 | |
| Deletion | Chr4 | 5842001 | 5868000 | 25999 | 6 |
| Deletion | Chr11 | 7859510 | 7859521 | 11 | |
| Deletion | Chr1 | 10780791 | 10780794 | 3 | |
| Deletion | Chr2 | 11489218 | 11489219 | 1 | |
| Deletion | Chr3 | 12103039 | 12103040 | 1 | |
| Deletion | Chr12 | 12484871 | 12484872 | 1 | |
| Deletion | Chr10 | 14054775 | 14054777 | 2 | |
| Deletion | Chr2 | 15858085 | 15858089 | 4 | |
| Deletion | Chr10 | 16016726 | 16016727 | 1 | |
| Deletion | Chr7 | 16913640 | 16913652 | 12 | |
| Deletion | Chr7 | 17187179 | 17187181 | 2 | |
| Deletion | Chr7 | 20135001 | 20227000 | 91999 | 11 |
| Deletion | Chr1 | 20144282 | 20144290 | 8 | |
| Deletion | Chr6 | 21421164 | 21421184 | 20 | |
| Deletion | Chr1 | 35637830 | 35637832 | 2 |
Insertions: 5
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 15151527 | 15458450 | LOC_Os02g25860 |
| Inversion | Chr2 | 15467472 | 15922963 | LOC_Os02g26330 |
| Inversion | Chr1 | 41335521 | 41558315 | 2 |
| Inversion | Chr1 | 41335522 | 41558316 | 2 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 7540638 | Chr2 | 15467448 | 2 |
| Translocation | Chr9 | 9009768 | Chr2 | 15461429 | |
| Translocation | Chr9 | 9357599 | Chr6 | 29946116 | 2 |
| Translocation | Chr8 | 25143134 | Chr2 | 24605938 |
