Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2387 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2387 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 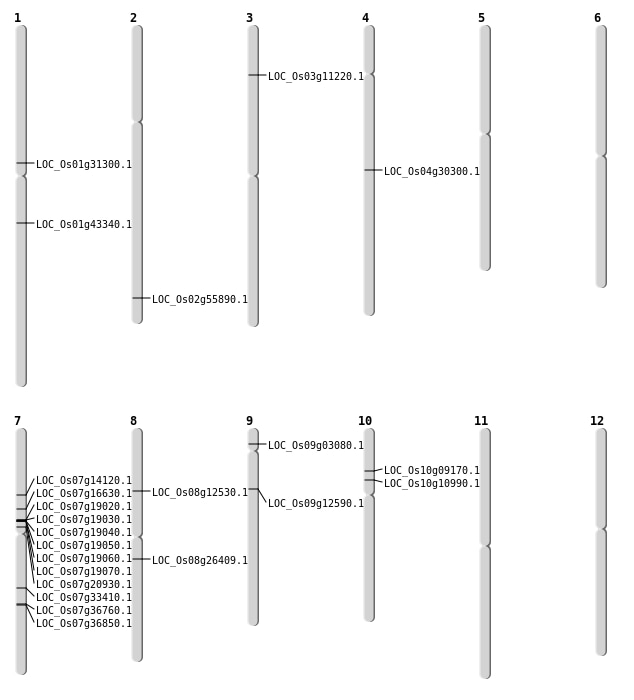
|
| Variant Information |
|---|
Single base substitutions: 53
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17123027 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g31300 |
| SBS | Chr1 | 2133394 | G-T | Homo | ||
| SBS | Chr1 | 23050847 | C-T | HET | ||
| SBS | Chr1 | 24774492 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g43340 |
| SBS | Chr1 | 26227059 | C-T | HET | ||
| SBS | Chr1 | 32570228 | A-G | Homo | ||
| SBS | Chr10 | 15903051 | C-G | HET | ||
| SBS | Chr10 | 16249881 | G-A | HET | ||
| SBS | Chr10 | 18510140 | G-A | HET | ||
| SBS | Chr10 | 4952446 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g09170 |
| SBS | Chr11 | 14107511 | G-A | Homo | ||
| SBS | Chr11 | 15062235 | G-T | HET | ||
| SBS | Chr11 | 1762023 | A-G | HET | ||
| SBS | Chr11 | 23769505 | C-T | HET | ||
| SBS | Chr11 | 5964650 | C-G | HET | ||
| SBS | Chr12 | 21820215 | C-T | HET | ||
| SBS | Chr2 | 14060854 | A-G | HET | ||
| SBS | Chr2 | 15305281 | G-A | HET | ||
| SBS | Chr2 | 19963858 | T-C | Homo | ||
| SBS | Chr2 | 20344539 | G-A | HET | ||
| SBS | Chr2 | 25898674 | T-C | HET | ||
| SBS | Chr2 | 33103216 | T-A | HET | ||
| SBS | Chr2 | 34213355 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g55890 |
| SBS | Chr2 | 4925482 | C-T | HET | ||
| SBS | Chr3 | 15680807 | T-A | HET | ||
| SBS | Chr3 | 15691324 | C-T | HET | ||
| SBS | Chr3 | 31041229 | G-A | HET | ||
| SBS | Chr3 | 3339287 | G-T | HET | ||
| SBS | Chr4 | 24171860 | G-A | Homo | ||
| SBS | Chr4 | 276502 | G-A | HET | ||
| SBS | Chr4 | 6780429 | G-T | HET | ||
| SBS | Chr5 | 17704500 | C-T | HET | ||
| SBS | Chr5 | 17744757 | A-G | HET | ||
| SBS | Chr5 | 19703803 | T-C | HET | ||
| SBS | Chr5 | 28437627 | G-A | HET | ||
| SBS | Chr5 | 9417364 | C-T | HET | ||
| SBS | Chr6 | 15511029 | C-T | Homo | ||
| SBS | Chr6 | 20124417 | G-A | HET | ||
| SBS | Chr6 | 31199284 | G-A | Homo | ||
| SBS | Chr6 | 5598312 | G-A | HET | ||
| SBS | Chr7 | 16387866 | G-A | HET | ||
| SBS | Chr7 | 19970779 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g33410 |
| SBS | Chr7 | 21814015 | C-T | HET | ||
| SBS | Chr7 | 22076001 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36850 |
| SBS | Chr7 | 4445179 | A-T | HET | ||
| SBS | Chr7 | 9765420 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g16630 |
| SBS | Chr8 | 16094165 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g26409 |
| SBS | Chr8 | 1959719 | G-A | HET | ||
| SBS | Chr8 | 27728768 | C-T | HET | ||
| SBS | Chr8 | 7416486 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g12530 |
| SBS | Chr8 | 7479418 | G-A | HET | ||
| SBS | Chr9 | 3033530 | A-G | HET | ||
| SBS | Chr9 | 8990173 | G-T | HET |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 1591415 | 1591447 | 32 | |
| Deletion | Chr6 | 1634233 | 1634235 | 2 | |
| Deletion | Chr6 | 3736733 | 3736734 | 1 | |
| Deletion | Chr11 | 3885041 | 3885044 | 3 | |
| Deletion | Chr5 | 5592337 | 5592338 | 1 | |
| Deletion | Chr3 | 5775224 | 5775232 | 8 | LOC_Os03g11220 |
| Deletion | Chr5 | 6917747 | 6917759 | 12 | |
| Deletion | Chr7 | 8059578 | 8059580 | 2 | LOC_Os07g14120 |
| Deletion | Chr11 | 8716549 | 8716554 | 5 | |
| Deletion | Chr7 | 11257001 | 11292000 | 34999 | 6 |
| Deletion | Chr10 | 13782479 | 13782482 | 3 | |
| Deletion | Chr7 | 13790086 | 13790091 | 5 | |
| Deletion | Chr6 | 14025204 | 14025214 | 10 | |
| Deletion | Chr6 | 15241734 | 15241735 | 1 | |
| Deletion | Chr10 | 15406979 | 15406988 | 9 | |
| Deletion | Chr4 | 18087918 | 18087923 | 5 | LOC_Os04g30300 |
| Deletion | Chr8 | 19335805 | 19335837 | 32 | |
| Deletion | Chr9 | 21987421 | 21987423 | 2 | |
| Deletion | Chr7 | 22024987 | 22024989 | 2 | LOC_Os07g36760 |
| Deletion | Chr7 | 22687142 | 22687143 | 1 | |
| Deletion | Chr7 | 26365363 | 26365365 | 2 | |
| Deletion | Chr2 | 32719718 | 32719729 | 11 |
Insertions: 6
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 6081079 | 6388849 | LOC_Os10g10990 |
| Inversion | Chr9 | 6553712 | 7219657 | 2 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 527336 | Chr9 | 1460113 | 2 |
| Translocation | Chr10 | 527342 | Chr9 | 1445245 | |
| Translocation | Chr8 | 16262952 | Chr7 | 12110042 | 2 |
