Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2426 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2426 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 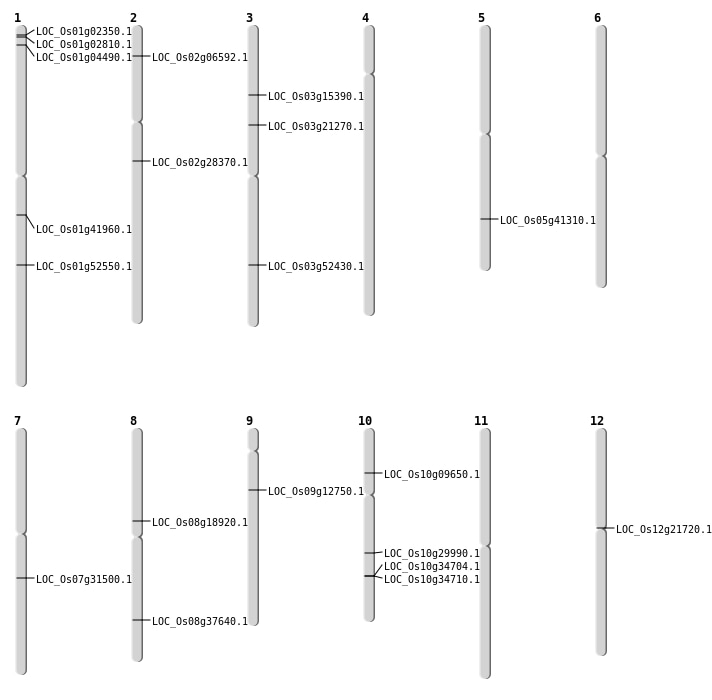
|
| Variant Information |
|---|
Single base substitutions: 58
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20036718 | A-G | HET | ||
| SBS | Chr1 | 2022604 | A-G | Homo | ||
| SBS | Chr1 | 23788103 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g41960 |
| SBS | Chr1 | 30190887 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g52550 |
| SBS | Chr1 | 35713900 | C-T | HET | ||
| SBS | Chr1 | 420151 | G-A | Homo | ||
| SBS | Chr10 | 14530471 | A-C | Homo | ||
| SBS | Chr10 | 5221391 | G-C | HET | SPLICE_SITE_ACCEPTOR | LOC_Os10g09650 |
| SBS | Chr10 | 5245832 | C-A | Homo | ||
| SBS | Chr10 | 5453929 | A-G | HET | ||
| SBS | Chr11 | 10567201 | C-A | Homo | ||
| SBS | Chr11 | 25168815 | T-C | Homo | ||
| SBS | Chr11 | 2625160 | G-T | HET | ||
| SBS | Chr12 | 12214269 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g21720 |
| SBS | Chr12 | 15349105 | C-T | HET | ||
| SBS | Chr12 | 22093380 | T-A | HET | ||
| SBS | Chr2 | 11098103 | T-C | HET | ||
| SBS | Chr2 | 29721856 | G-A | HET | ||
| SBS | Chr2 | 3314322 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g06592 |
| SBS | Chr2 | 4866184 | C-A | HET | ||
| SBS | Chr3 | 12153270 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g21270 |
| SBS | Chr3 | 23199603 | G-A | Homo | ||
| SBS | Chr3 | 23877152 | G-T | HET | ||
| SBS | Chr3 | 30086641 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g52430 |
| SBS | Chr3 | 30719103 | C-T | Homo | ||
| SBS | Chr3 | 30719112 | C-T | Homo | ||
| SBS | Chr3 | 31370257 | T-A | Homo | ||
| SBS | Chr4 | 18314082 | A-C | HET | ||
| SBS | Chr4 | 4909651 | G-A | HET | ||
| SBS | Chr5 | 13262739 | C-A | HET | ||
| SBS | Chr5 | 14576811 | A-G | HET | ||
| SBS | Chr5 | 20173072 | A-C | Homo | ||
| SBS | Chr5 | 20614536 | C-T | Homo | ||
| SBS | Chr5 | 24197520 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g41310 |
| SBS | Chr5 | 26664587 | A-G | HET | ||
| SBS | Chr5 | 29928254 | G-T | Homo | ||
| SBS | Chr6 | 13957721 | G-T | HET | ||
| SBS | Chr6 | 15093295 | G-A | HET | ||
| SBS | Chr6 | 15775256 | A-G | HET | ||
| SBS | Chr6 | 24875707 | G-A | HET | ||
| SBS | Chr6 | 27078473 | C-T | HET | ||
| SBS | Chr6 | 3386347 | G-T | HET | ||
| SBS | Chr7 | 14897830 | G-A | HET | ||
| SBS | Chr7 | 18690226 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g31500 |
| SBS | Chr7 | 27053370 | A-C | HET | ||
| SBS | Chr7 | 28812390 | G-A | HET | ||
| SBS | Chr7 | 29618800 | T-C | HET | ||
| SBS | Chr7 | 8039264 | T-A | HET | ||
| SBS | Chr7 | 8074851 | C-T | HET | ||
| SBS | Chr7 | 9431632 | T-C | HET | ||
| SBS | Chr8 | 11293455 | T-A | HET | ||
| SBS | Chr8 | 23269838 | T-C | HET | ||
| SBS | Chr8 | 24982424 | A-G | HET | ||
| SBS | Chr8 | 7294163 | G-A | HET | ||
| SBS | Chr8 | 777235 | T-A | Homo | ||
| SBS | Chr9 | 14623894 | C-G | HET | ||
| SBS | Chr9 | 19230192 | C-A | HET | ||
| SBS | Chr9 | 7321733 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g12750 |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 743784 | 743788 | 4 | LOC_Os01g02350 |
| Deletion | Chr1 | 988208 | 988215 | 7 | LOC_Os01g02810 |
| Deletion | Chr11 | 1699387 | 1699391 | 4 | |
| Deletion | Chr8 | 2629959 | 2629960 | 1 | |
| Deletion | Chr6 | 3102684 | 3102685 | 1 | |
| Deletion | Chr3 | 3768404 | 3768410 | 6 | |
| Deletion | Chr6 | 6262001 | 6273000 | 10999 | |
| Deletion | Chr4 | 6707698 | 6707718 | 20 | |
| Deletion | Chr3 | 7133862 | 7133869 | 7 | |
| Deletion | Chr3 | 8418482 | 8418494 | 12 | LOC_Os03g15390 |
| Deletion | Chr12 | 10127262 | 10127264 | 2 | |
| Deletion | Chr8 | 11277591 | 11277593 | 2 | LOC_Os08g18920 |
| Deletion | Chr8 | 11344149 | 11344154 | 5 | |
| Deletion | Chr5 | 13919748 | 13919749 | 1 | |
| Deletion | Chr5 | 14467657 | 14467679 | 22 | |
| Deletion | Chr9 | 15212219 | 15212237 | 18 | |
| Deletion | Chr6 | 16376963 | 16376964 | 1 | |
| Deletion | Chr2 | 16767646 | 16767654 | 8 | LOC_Os02g28370 |
| Deletion | Chr10 | 18512001 | 18538000 | 25999 | 2 |
| Deletion | Chr5 | 21724129 | 21724142 | 13 | |
| Deletion | Chr8 | 23849963 | 23849966 | 3 | LOC_Os08g37640 |
| Deletion | Chr1 | 25496407 | 25496418 | 11 | |
| Deletion | Chr6 | 28809991 | 28810001 | 10 | |
| Deletion | Chr1 | 33766350 | 33766381 | 31 | |
| Deletion | Chr3 | 35701989 | 35701990 | 1 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr6 | 24414618 | 24414619 | 2 | |
| Insertion | Chr8 | 3946507 | 3946507 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 2000541 | 2117307 | LOC_Os01g04490 |
| Inversion | Chr1 | 2000549 | 2117319 | LOC_Os01g04490 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 23626982 | Chr10 | 15566640 | 2 |
| Translocation | Chr12 | 23626985 | Chr10 | 15450841 |
